RNAi Screening Trends

Complete the form below to unlock access to ALL audio articles.
Introduction
RNA-mediated interference (RNAi) is standard tool for sequence-specific gene knockdowns which are typically associated with a phenotypic change and can lead to the identification of unknown components of biological pathways. RNAi has been around for about a decade and enable high-throughput loss-of-function screens against the human genome. The prospective researcher needs to make a number of decisions before proceeding with RNAi screening. These include deciding whether to screen for targets individually (so-called ‘arrayed’ approach) or do pooled RNAi screens; choosing whether to screen genome wide or to focus on subsets of already annotated gene families (e.g. target-class specific); and identifying the best experimental system and type of screen with which to answer the biological question of interest. Some of the technical considerations include the method of RNAi delivery and endpoint monitored for detecting the desired phenotype robustly, reliably and in high-throughput format. The RNAi screener also needs an awareness of recent developments in clustered regularly interspaced palindromic repeats (CRISPR) technology, as there are situations where the CRISPR approach to genome editing may be more appropriate than RNAi.
In October 2014 HTStec undertook a market survey on RNA interference (RNAi) screening and gene silencing mainly among research labs in academia, pharma and biotech. The main objectives were to comprehensively document current practices and preferences in RNAi screening, and to understand future user requirements. This article contains ‘selected findings’ from the HTStec market report ‘RNA Interference Screening Trends 2014’. It is intended to provide the reader with a brief insight into recent market trends. It covers only 10 out of the 30 original questions detailed in the full report. The full published report should be consulted to view the entire dataset, details of the breakdown of the response to each question, its segmentation and the estimates for the future. Please click here for more information.
Disease areas targeted by RNAi screening:
The key disease or research area most targeted with RNAi screening assays are presented in Figure 1. This showed that the oncology/cancer area was most targeted (63% targeting). This was followed by immunology/inflammatory disease/autoimmune (33% targeting); infectious diseases (31% targeting); and then neurology/CNS/ neurodegeneration/ pain and metabolic disease/diabetes (both 24% targeting).

Figure 1 Key Diseases Or Research Areas Targeted With RNAi Screening Assays
Assay preferences when undertaking RNAi screens:
The main application areas of RNAi screening assays are reported in Figure 2. This showed that the identification of new therapeutic targets was the main application of RNAi screening (57% applying). This was followed by unbiased screens to identify genes that regulate various cell biological processes (47% applying); screens to identify novels parts of signalling pathways/cascades (45% applying); and then screens to identify genes that regulate cancer (41% applying). Of least interest was RNAi therapeutic technology development (only 20% applying).
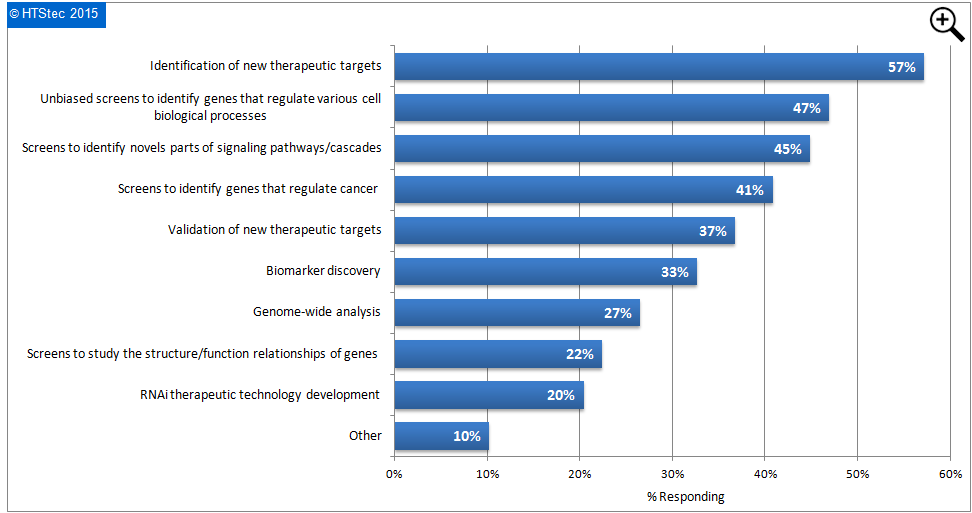
Figure 2 Main Application Areas For RNAi Library Screening
The use of the two main approaches to RNAi screening is given in Figure 3. i.e. 1) Arrayed where each unique RNAi reagent or unique set of reagents targeting a single gene, such as for a small pool of independent siRNAs targeting one gene, occupies a unique well in a microtiter plate, such as a 384-well plate; versus 2) Pooled where all of the gene-specific reagents are pooled together, or synthesized en masse, and added, at random, to cells. This showed that survey respondents primarily used an arrayed approach (46% preferring), or they used both an arrayed and a pooled approach (42% preferring), leaving only 13% who preferred primarily a pooled approach.

Figure 3 Preferred Approach To RNAi Screening
The main types of RNAi screens investigated today (2014) are presented in Figure 4. This showed the most investigated were loss of function screens - based on decrease or loss of a phenotype (65% using). This was closely followed by loss of function screens - based on enhancement of a phenotype (54% using); and then synthetic lethal screens - based on dual knock-downs or parallel knock-out cell lines to create a synthetic lethality (28% using). The use of CRISPR reversible knockdowns was at present limited to only a few respondents (7% using).

Figure 4 Main Types of RNAi Screens Investigated Today (2014)
The main endpoints (readouts) monitored when undertaking RNAi screening are reported in Figure 5. This showed that either plate reader based monitoring of whole cell responses (e.g. viability, cell proliferation) or high content microscopic imaging of phenotypic markers/cellular parameters were the most used endpoints (both with 62% using). This was followed by quantification of mRNA by qRT-PCR (40% using); plate reader based cellular imaging combined with bulk monitoring of whole cell response (21% using); and then high content laser scanning cytometry of phenotypic markers/cellular parameters (19% using).

Figure 5 Main Endpoints Monitored When Undertaking RNAi Screening
Transfection methods used in siRNA library screening:
The delivery (transfection) methods most used today (2014) for siRNA library screening were ranked in Figure 6. This showed that standard chemical (lipid-mediated) transfection ranked as most used. This was followed by reverse transfection and then lipid-free transfection. Least used today were electroporation and viral transfection.
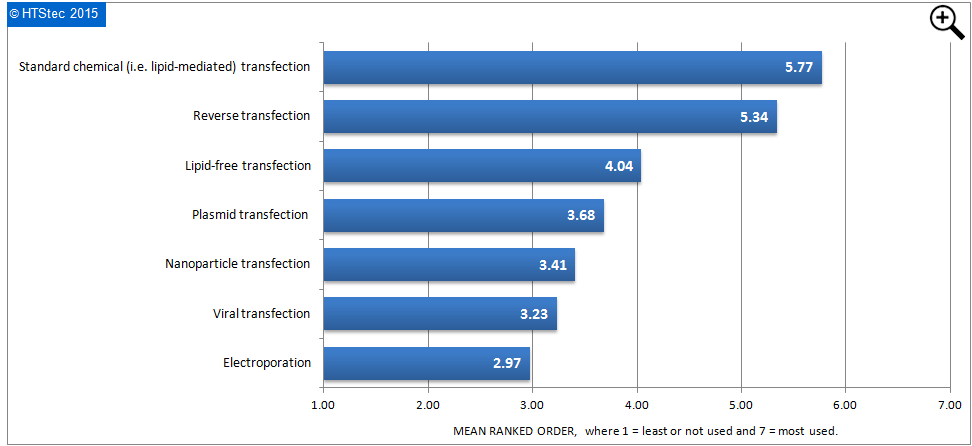
Figure 6 Delivery (Transfection) Methods Most Used Today In siRNA Library Screening
RNAi library screening:
The vendor compiled RNAi libraries of greatest interest to survey respondents are reported in Figure 7. This showed that those RNAi libraries covering the Human Genome were of most interest (33% preferring). This was followed by target class specific libraries e.g. kinases, proteases, GPCR, ion channels etc. (28% preferring) and then pathway analysis specific (20% preferring).

Figure 7 Vendor Compiled RNAi Libraries Of Greatest Interest To Respondents
Biggest challenges in RNAi screening:
What most limits survey respondent’s work on RNAi screening today is ranked in Figure 8. This showed that off-target effects were ranked most limiting. This was closely followed by the level of knock-down; knock-down does not produce a robust phenotypic change; and then post-screen analysis methods/deconvolution. Ranked least limiting was inadequate control strategies.
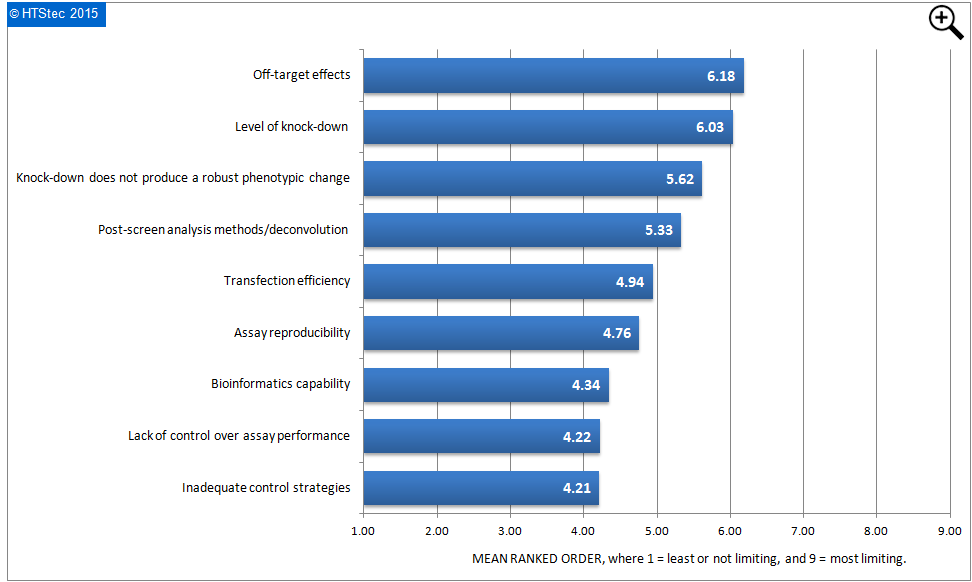
Figure 8 What Most Limits Respondent's Work On RNAi Screening Today
Vendor awareness in supplying RNAi screening reagents:
The supplier/vendor of RNAi screening reagents and consumables that first comes into the mind of survey respondents is presented in Figure 9. This showed that Thermo Scientific/Life Technologies was the most recognized vendor of RNAi screening reagents and consumables (with 38% selecting). This was followed by GE Dharmacon (26% selecting); Sigma Aldrich/Proligo (13% selecting); Qiagen (8% selecting); and then EMD-Millipore (5% selecting). All others had a combined total of less than10% of all selections. Please note this should NOT be regarded as a true market share projection. It is based on the supplier of RNAi screening reagents and consumables that first comes into the mind of survey respondents, which may not be the same as those who they purchase most consumables from.

Figure 9 Supplier/Vendor Of RNAi Screening Reagents & Consumables That First Comes To Mind
Preferred gene silencing approach:
The discovery that the CRISPR-Cas9 bacterial immune system can be re-purposed to easily create deletions, insertions and replacements in the mammalian genome has the potential to revolutionize the field of genome engineering and re-invigorate the field of gene therapy. CRISPR disrupts a gene when a mistake occurs in the repair of a DNA break it introduces in the targeted gene. RNAi disrupts gene function in a very different manner, it interferes with the translation of genes by inducing rapid degradation of a gene target's transcript. RNAi may therefore be preferable to CRISPR for validating new drug targets because small molecule drugs only reduce a gene’s function somewhat, as RNAi does, rather than knocking it out entirely, as CRISPR does. There is currently high interest in using CRISPR as an alternative approach to RNAi in gene silencing. With this in mind survey respondents were asked, based on their existing knowledge, which approach they currently perceive to be most advantageous with respect to a list of desired attributes of a technology used for gene silencing. These results are presented in Figure 10. This showed that RNAi approaches were perceived more advantageous by most survey respondents for ease of use, broad applicability, short experimental turnaround, and cost-effectiveness. In contrast CRISPR approaches were regarded as preferable only for specificity; and good interpretability of results.
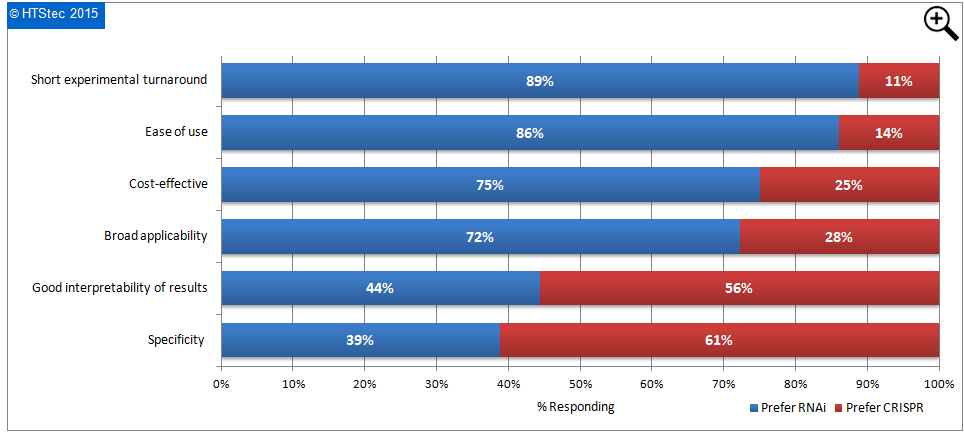
Figure 10 Gene Silencing Approach (RNAi versus CRISPR) Perceived To Be Most Advantageous With Respect To Desirable Technology Attributes
These findings are not in alignment with some of the advantages of CRISPR put out by the reagent providers and would suggest more needs to be done the clarify the advantages of CRISPR versus RNAi.
Conclusions:
The selected findings reported above revealed that oncology/cancer was the disease or research area most targeted with RNAi screening assays. The main application area of RNAi screening assays was the identification of new therapeutic targets. Most RNAi screening performed today involved an arrayed approach where each unique RNAi reagent or unique set of reagents targeting a single gene, (such as for a small pool of independent siRNAs targeting one gene) occupies a unique well in a microtiter plate. The main types of RNAi screens investigated today were loss of function screens either based on decrease or loss of a phenotype or based on an enhancement of a phenotype. The main endpoints (readouts) monitored when undertaking RNAi screening were either plate-reader based monitoring of whole cell responses e.g. viability, cell proliferation or high content microscopic imaging of phenotypic markers/cellular parameters. The delivery (transfection) methods most used today for siRNA library screening was standard chemical (i.e. lipid-mediated) transfection. The vendor compiled RNAi libraries of greatest interest to survey respondents were those covering the entire Human Genome.
These selected findings also confirmed that one of the biggest challenges of RNAi screening today was off-target effects, i.e. most siRNAs not only target their complementary on-target gene but also deregulate an unknown number of off-target genes. These off-target effects are sequence-specific and intrinsic to each siRNA. Even though some progress has been made towards reducing off-target effects, most available RNAi reagents do not yet provide satisfactory protection. Some of the new siPOOLs from siTOOls Biotech, could provide promise in this respect.
The suppliers/vendors of RNAi screening reagents and consumables that first came into the mind of survey respondents were Thermo Scientific/Life Technologies, GE Dharmacon and Sigma Aldrich/Proligo. All three of these vendors are at the forefront of delivering the broadest range of gene editing technologies, including the CRISPR-Cas9 system.
Survey respondents currently perceived RNAi approaches for gene silencing were technologically more advantageous than CRISPR for ease of use; broad applicability; short experimental turnaround; and cost-effective, whilst CRISPR interference approaches were regarded as preferable only for specificity; and good interpretability of results. Recent findings suggests that RNAi and the CRISPR-Cas9 system are complementary technologies with good overlap and that they can be used to validate each other’s results. With both technologies, researchers now have an impressive toolbox for elucidating gene function and enabling greater understanding of the genome in relation to drug discovery.
DISCLAIMER: HTStec Limited has exercised due care in compiling and preparing these Selected Findings from its Report, which is based on information submitted by individuals in respondent companies. HTStec Limited has NOT verified the accuracy of this information, nor has it is established respondent’s authority to disclose information to HTStec Limited. HTStec Limited expressly disclaims any and all warranties concerning these Selected Findings including any warranties of mechantability and/or fitness for any particular purpose, and warranties of performance, and any warranty that might otherwise arise from course of dealing or usage of trade. No warranty is either expressed or implied with respect to the use of these Selected Findings. Under no circumstances shall HTStec Limited be liable for incidental, special, indirect, direct or consequential damages or loss of profits, interruption of business, or related expenses that may arise from use of these Selected Findings, including but not limited to those resulting from inaccuracy of the data therein.

