Visualising the Problem may help to Improve Antibiotics

Complete the form below to unlock access to ALL audio articles.
Professor William Hunter, from the College of Life Sciences at Dundee, coordinates the EU-funded Aeropath project, which is designed to advance early stage drug discovery against Pseudomonas aeruginosa (P. aeruginosa), a common bacterium that can cause serious diseases in animals and humans.
This bacterium is particularly troublesome since it has developed resistance to many common antibiotics and is an important pathogen of burns victims, immuno-compromised patients for example due to chemotherapy or other conditions, and also to cystic fibrosis patients.
In collaboration with colleagues from Oxford’s Division of Structural Biology, and the Oxford Protein Production Facility (OPPF) using state of the art facilities at the Diamond Light Source synchrotron in Oxfordshire, the team were able to determine the accurate chemical structure of the penicillin binding protein PBP3 from Pseudomonas aeruginosa.
The research was made possible by using a machine called a synchrotron. It houses a large ring-shaped vacuum tube similar in size to Hampden Park, in which electrons are accelerated at close to the speed of light and manipulated by special magnets to give off very intense X-rays. These X-rays were then used to probe tiny PBP3 crystals through diffraction methods, enabling the researchers to determine the 3D structure of the protein.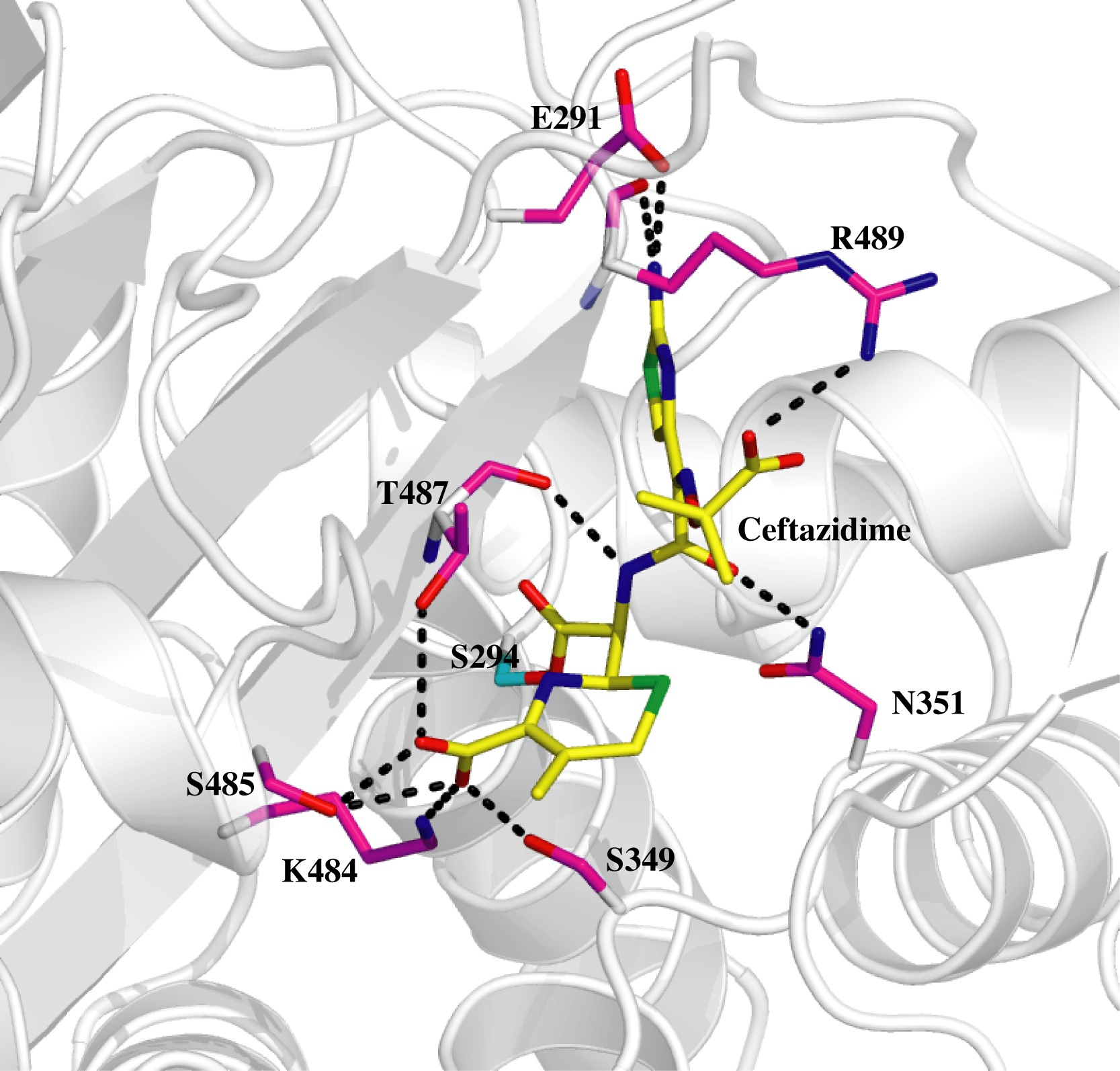
The research has been published in the most recent edition of the Journal of Molecular Biology. Now that the Dundee/Oxford team have established the exact chemical structure of the protein, researchers at these and other institutions will be able to try and develop new inhibitors and therapies.
Using the Diamond synchrotron the team were able to efficiently determine the 3D structure of the penicillin binding protein PBP3, and this may have significant impact in the future development of antibiotics, according to Professor Hunter.
“Having this accurate 3D picture of the enzyme and knowing where these molecules called inhibitors bind is critical,” he said. “It gives us a clear understanding of the molecular interactions that are involved in inhibiting this drug target. If we didn’t have that information we would be looking to characterise interactions by other less accurate methods.
“In those circumstances, you’re essentially working blind. You have to do many more experiments to get the understanding we have been able to get by doing this crystal structure.
“The objective of our work is to help advance understanding of potentially new targets for the development of therapies against Gram negative infections, which are a real problem because of the increase in drug resistance and in some cases, these are just tough beasts to kill.
“Pseudomonas aeruginosa is something we’re particularly interested in as this causes a lot of problems for people whose immune system is compromised either due to transplant surgery, chemotherapy, people who have HIV/AIDS, people who are recovering from burns and also young people with cystic fibrosis whose lungs struggle to cope with dangerous Gram negative bacteria.
“Because these organisms are so tough, we need new ideas for drugs and the way to do that is to find new targets or exploit old targets and come out with new compounds that will hit the old targets.
“We interact with a number of colleagues to look at these targets. In order to make sure we were casting our net as wide as possible I set up a collaboration with this group in Oxford to get them to take on additional targets and, using the excellent facilities at the Diamond synchrotron, they were fantastic and did a very good job of getting the structure of the penicillin binding protein PBP3.
“For many years, this has been a key therapeutic target but the organisms can change, mutate, and develop drug resistance.
“Now that we’ve got this 3D picture, we know where all the atoms are, we know how this molecule works, and the challenge is to use this information to come up with new small molecules that will stop PBP3 from working. This is a medical problem that deserves a solution and we are looking at several possible pathways of modifying existing drugs to supplement the arsenal of antibiotics.”
The team used Diamond Light Source’s protein crystallography facilities to solve three structures: the protein in native form; and the protein linked to two important antibiotics, carbenicillin and ceftazidime.
A common feature observed in the crystal structures of PBPs is the flexibility of the active sites. This property plays a key role in the bacteria’s ability to develop a resistance to drugs; the resistance mechanism works by mutating the protein. For binding of the drug to occur, the protein bends towards the inhibitor. The conformational change introduced by the inhibitor opens up a small pocket within the active site.
From their 3D structures of the mechanism, the group are able to characterise these pockets with a view to targeting them with a new type of inhibitor that restricts the active site “breathing” i.e. restrict the flexibility of the protein. This new inhibitor could be used in conjunction with existing antibiotics to combat drug resistance.
Overall, the group’s findings will allow discussions of different approaches to inhibit PBPs and potentially lead to new ways of combating P. aeruginosa and related bacteria which cause respiratory diseases and skin infections, gastro-intestinal and sexually transmitted diseases.
The Dundee team is funded by the European Commission, while the Medical Research Council (MRC) provide funding for the Oxford researchers’ work.

