4 Ways To Reduce Non-specific Binding in Surface Plasmon Resonance Experiments

Complete the form below to unlock access to ALL audio articles.
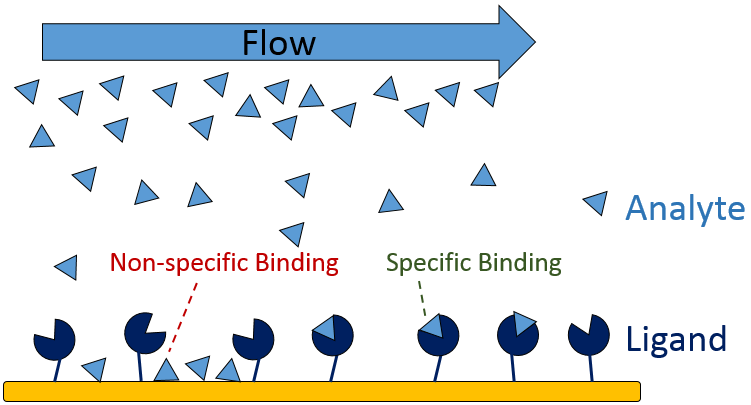
SPR experiments typically consist of a ligand, the biomolecule that is coupled to the sensor, and the analyte, the biomolecule that binds to the ligand. Sometimes the analyte interacts non-specifically with the sensor chip as well. In the SPR world, this is called non-specific binding (NSB), as the purpose of SPR experiments is to measure only the interaction between the analyte and the ligand, and not the background. NSB is caused by molecular forces (charge interactions, hydrophobic interactions, hydrogen bonding, etc) between the analyte and the sensor surface. In order to test for NSB, a preliminary binding test with only the bare sensor (sensor without the ligand) and the analyte should be completed. If there is significant response when the analyte is passed over the bare sensor (false positive), do not stress, as there are a variety of steps you can take to reduce NSB for SPR experiments. Before you get started, it is important to know the profile of your analyte, for example, the isoelectric point, charge, size and the composition (hydrophilic, hydrophobic), so you can choose the best method possible for reducing NSB. The most common methods to prevent NSB can be found below.

Example of the reduction of charge non-specific binding with the addition of 200 mM NaCl. Red curves represent non-specific binding from various analyte concentrations. Black curves represent the corresponding response of analyte with the addition of salt to running buffer. For a detailed experimental analysis of these methods, download our technote.
1) Adjust your pH
The pH of the buffer you are using for SPR experiments can have a significant impact on non-specific binding because it dictates whether your biomolecule has an overall positive or negative charge. For example, if you are using a pH at which your protein (analyte) is positively charged and the sensor surface is negatively charged, you can definitely expect to see non-specific charge interactions. You can fix this problem by preparing a buffer with a pH within the isoelectric point range (predicted neutral overall charge) of your protein.
2) Use Additives – Protein Blocker
If you are using a protein as your analyte, you can always try using bovine serum albumin (BSA) in your buffer and samples to help prevent non-specific binding. BSA is a globular protein with hydrophilic and hydrophobic subgroups which can surround your protein to protect it from binding to other proteins, charged surfaces, glass and plastic. Try to keep your BSA concentration at 1% or lower. Additives such as BSA and tween are used in most experiments not only to prevent non-specific binding but also to prevent analyte loss to the tubing walls at low concentrations.
3) Use Surfactants – Tween 20
In non-specific binding occurring due to hydrophobic interactions in your system? Low concentrations of mild detergents such as Tween 20 can disrupt hydrophobic interactions between your analyte and sensor. Tween 20 is commonly added to biological systems to prevent the analyte from binding to tubing and containers as well.
4) Increase salt concentration (NaCl)
Using higher salt concentrations in your buffer and sample can reduce non-specific binding due to charge interactions by producing a shielding effect. Salts such as NaCl prevent charges on the protein from interacting with charges on the surface. For example, adding 200 mM NaCl to the running buffer and samples can disrupt the charge interactions between a protein and carboxyl surface to reduce NSB binding completely.
Preventing non-specific binding (if it exists) is key when performing surface plasmon resonance experiments. The more you know about the biological constituents of your system, the easier optimizing for NSB will be. It is also important to note that certain extreme conditions might deactivate or denature your biomolecules. For example, NSB might be completely eliminated at pH 10, but your protein might denature under those conditions, so you would have to adjust the conditions further. However, the complete elimination of non-specific binding is not always necessary if the quantity of specific binding is much greater than the signal of NSB.
by Tijana Matovic, Nicoya Lifesciences

