Color-Coded Stem Cells

Complete the form below to unlock access to ALL audio articles.
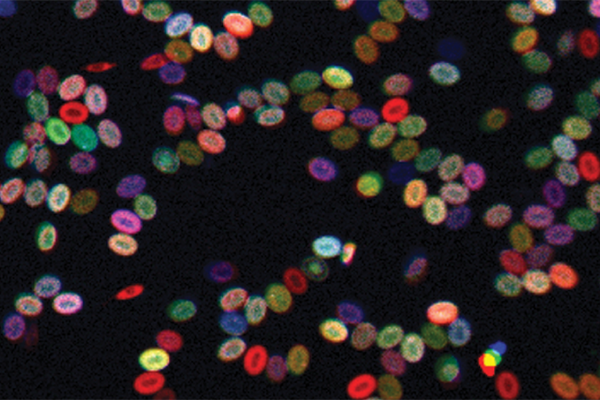
A new color-coding tool is enabling scientists to better track live blood stem cells over time, a key part of understanding how blood disorders and such cancers as leukemia arise, report researchers at Harvard Medical School and Boston Children’s Hospital.
In Nature Cell Biology on Nov. 21, they describe the use of their tool in zebrafish to track the blood stem cells the fish are born with, the clones these cells make of themselves and the types of specialized blood cells they give rise to (red cells, white cells and platelets).
Leonard Zon, professor of stem cell and regenerative biology in the Faculty of Arts and Sciences at Harvard University and a senior author on the paper, believes the tool has many implications for hematology and cancer medicine since zebrafish are surprisingly similar to humans genetically.
People are born with a certain number of blood stem cells and rely on them for life. Various blood disorders and cancers are thought to arise when a mutant clone of an original blood stem cell starts to dominate. But what actually happens with blood stem cells over time has been hard to pin down.
“There’s significant interest in determining how a stem cell clone expands, what makes one clone dominant, and why that predisposes you to cancer and blood disorders,” said Zon, who is also director of the Stem Cell Research Program at Boston Children’s.
All the colors of the rainbow
Zon, with David Traver of the University of California at San Diego, first author Jonathan Henninger of HMS and Boston Children’s, and other colleagues, used a specially bred zebrafish called Zebrabow that has multiple copies of genes for red-blue-green fluorescent protein scattered through its genome.
By activating certain enzymes, the team cut the red-blue-green assembly in different ways and in different amounts. This technique yields, in theory, about 80 different colors based on the cell’s overall proportions of each fluorescent protein. Each color represents a different clone or variety of blood stem cell.
“It’s like an RGB television set, where red, blue and green give you the whole spectrum of colors,” explained Zon. “In our system, the enzyme cuts out different parts—blue, for example, or green and blue—so the stem cell will end up a different shade of color. In this way, we were able to mark each stem cell being born with a different color, and then follow the colors through development and see how many stem cells of each color were present in the adult fish.”
Other scientists have developed tracking systems based on genetic barcodes. But these require dissecting the cells, so cannot analyze living, circulating cell populations.
“Because our system is based on color, we don’t have to destroy the cells to analyze their clonality,” said Henninger, an HMS research assistant graduate student in the Zon lab. “Rather, we can isolate the cells by color, with the help of computational tools, and then look at what genetic factors are involved in their expansion.”
Tracking blood disease and cancer
A normal blood system tends to have a diversity of clonal stem cell types; all cells are genetically identical but have different epigenetic changes that affect gene expression. Being able to track how different clonal populations develop has many implications for medicine.
For example, leukemia and myelodysplastic disorders, in which certain kinds of blood cells aren’t made or don’t mature properly, are thought to arise from a clone of an original stem cell that ran amok and began replicating in great numbers.
But until now, this has been hard to analyze, since no one knew how many blood stem cells we start out with. Based on the zebrafish data, Henninger and Zon estimate that blood stem cells make up about 20 percent of all blood cell progenitors at the time they are formed.
That provides a starting point for exploring, for example, why and how a particular blood stem cell clone may begin to expand as people age, posing a risk for leukemia, or how cancer chemotherapy can sometimes transform tumor cells.
“Within a tumor, you can get clones of cells that become resistant to chemotherapy, and that’s what kills the person,” Zon explained. “The questions are, ‘What makes a clone take off and start to behave differently than the rest? How is clonal diversity regulated?’”
Maximizing marrow transplant
Similarly, the knowledge could also improve bone marrow transplantation for a variety of childhood and adult disorders.
“When you do a bone marrow transplant, only certain clones of stem cells take,” Zon said. “This is important because these clones are driving development of an entire blood system, based on a finite number of cells.”
To test this out, the team irradiated the Zebrabow fish, damaging their blood cells, then watched, using their color-coding system, as the fish’s blood systems recovered.
“We saw that a smaller number of clones were dominant,” Zon said. “This suggests there are some ‘good’ cells that help people regenerate a healthy blood system.”
Source:
Story from Harvard Medical School. Original piece written by Nancy Fliesler. Please note: The content above may have been edited to ensure it is in keeping with Technology Networks’ style and length guidelines.
Reference:
Henninger, J., Santoso, B., Hans, S., Durand, E., Moore, J., Mosimann, C., … Zon, L. (2016). Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nature Cell Biology. doi:10.1038/ncb3444

