Making Metabolite Identification More Efficient

Complete the form below to unlock access to ALL audio articles.
MetID and DMPK groups from a variety of industries have struggled with the same challenges when it comes to metabolite identification. Specifically, the amount of time required to prepare data before spectral interpretation, the verification of detected and identified metabolites, the compilation of interpreted data, the creation of reports, and finally, the difficulties with disseminating information and knowledge due to disparate data storage. Overall, scientists have lacked the tools that would eliminate the burden of manually analyzing data. Using a solution that automates metabolite identification allows scientists to be more efficient with more comprehensive results from a combined prediction and data-driven approach.
MetaSense offers scientists a significant advantage over existing methodologies with the ability to yield faster, easier and more accurate detection and identification of predicted and unexpected metabolites. The solution combines and interprets LC/MS analytical data from all instrument vendors, reducing bottlenecks and efficiently generating biotransformation maps. The solution also produces answers at a faster rate that are shared in a centralized location with assembled live spectra, chromatograms, and structures. Lastly, MetaSense facilitates decision-support and effective knowledge sharing throughout organizations and between partners.
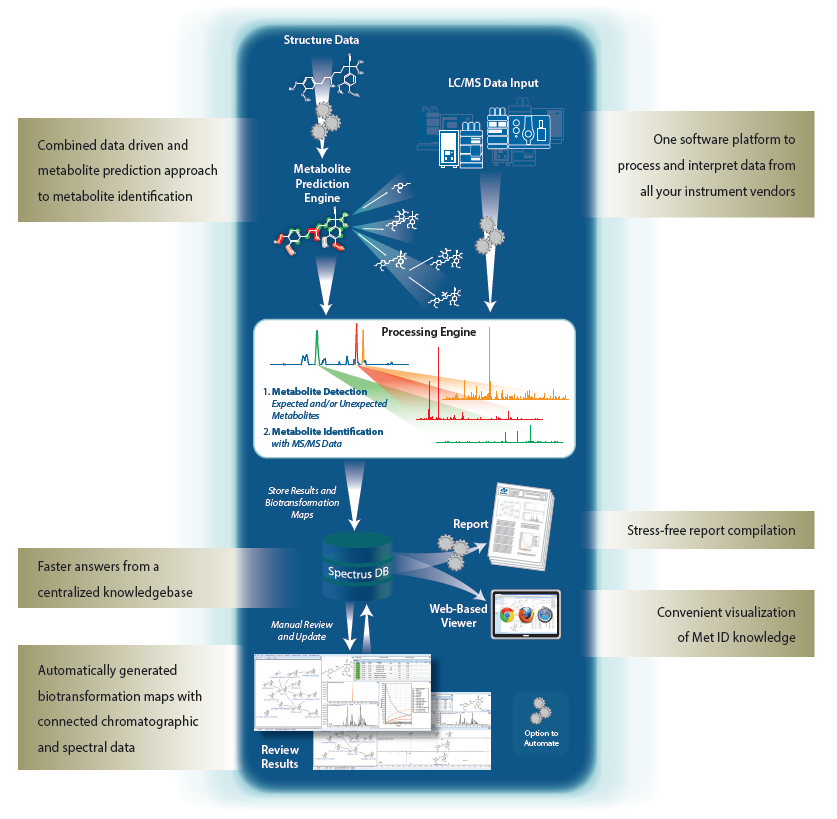
Built on the ACD/Spectrus Platform, MetaSense offers an array of functionality. To learn more, visit: the further information link below.



