The Ras Pathway and Cancer: Regulation, Challenges and Therapeutic Progress

Complete the form below to unlock access to ALL audio articles.
For many years, the Ras signaling pathway has triggered both hope and frustration in the field of cancer research. In normal health, Ras signaling is central to the regulation of intracellular signaling networks involved in cell proliferation, growth and survival. Like many cellular signaling pathways, however, the Ras-related cascade has a dark side. Mutations in the RAS gene itself, or in upstream or downstream regulators, can push the pathway into a constitutively active, cancer-inducing state.
In a recent analysis of major cancer mutation databases, it was estimated that approximately 19% of patients with cancer harbor RAS mutations – the equivalent of 3.4 million new cancer cases each year, globally.1 Therefore, finding ways to inhibit aberrant Ras signaling remains a key research priority.2
What is the Ras signaling pathway?
H-Ras, K-Ras and N-Ras are closely related proteins that are frequently mutated in human cancers. Specifically, they are different isoforms of small, guanosine triphosphatases (GTPases) that function as binary switches in all mammalian cells. Normally, Ras proteins are tightly controled, flicking between an active (bound to guanosine triphosphate, GTP) and inactive state (bound to guanosine diphosphate, GDP). When turned “on” they trigger a complex signaling cascade that transduces cell survival and growth signals.3 While Ras functions are regulated on many levels, the core pathway involves Ras, Raf (rapidly accelerated fibrosarcoma), MEK (mitogen-activated extracellular signal-regulated kinase) and ERK1/2 (extracellular signal-related kinase 1 and 2). Commonly referred to as the Ras-Raf-MEK-ERK cascade (Figure 1), it is the central, evolutionarily conserved pathway that transmits signals from cell surface receptors to transcription factor targets downstream via the following sequence:3
- Binding of epidermal growth factor (EGF) to the EGF receptor activates a guanine exchange factor (e.g., SOS) to load Ras with GTP.
- Ras-GTP recruits Rafs or Raf/MEK heterodimers to the plasma membrane.
- Raf-MEK assemblies activate RAF.
- Active Raf dimers activate MEK.
- Activated MEKs phosphorylate ERKs (a subgroup of mitogen-activated protein kinases, MAPK).
- ERK phosphorylates diverse substrates involved in cell proliferation, survival, growth, metabolism, migration and differentiation.
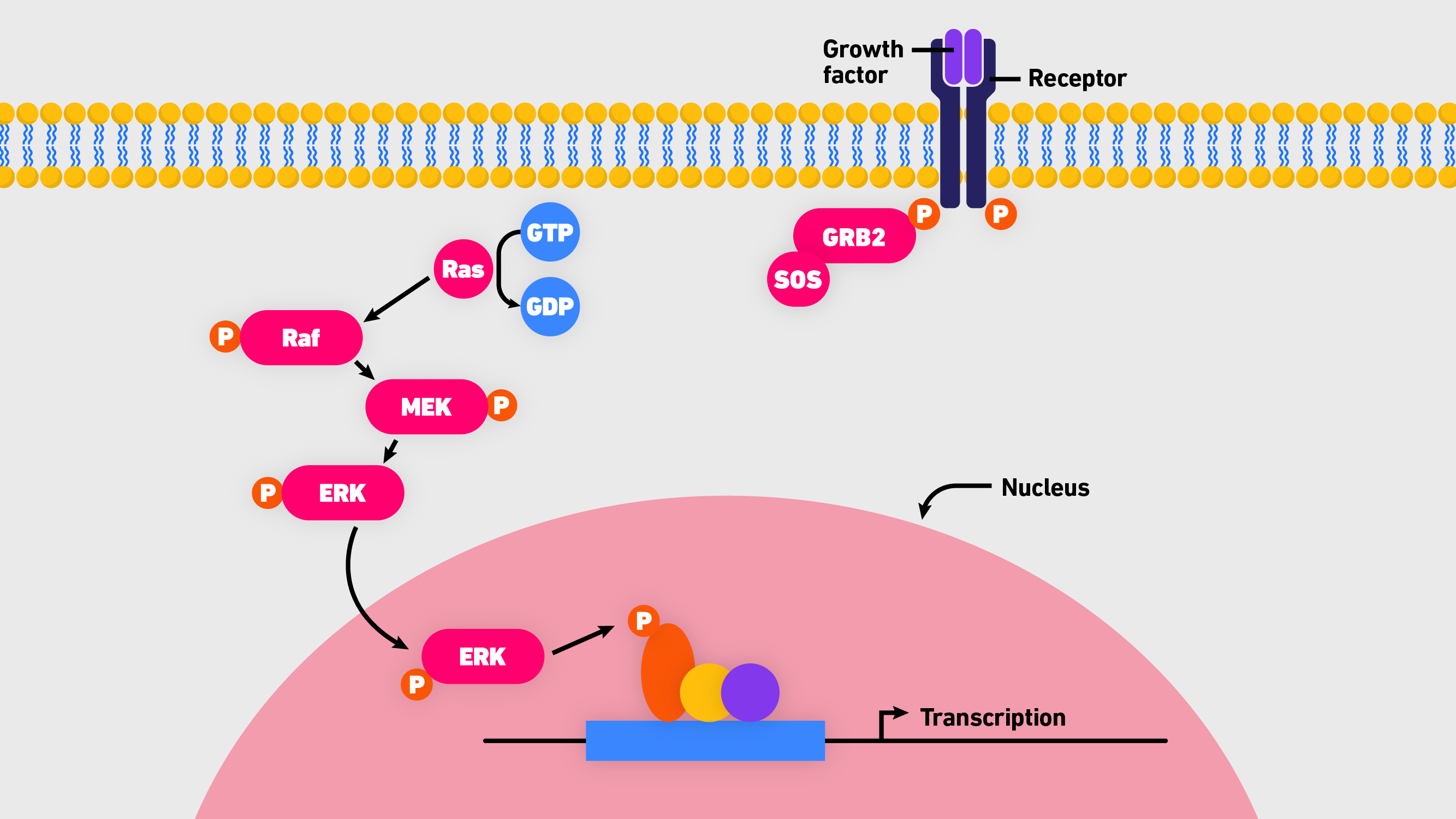
Figure 1: An overview of the Ras-Raf-MEK-ERK cascade.
The Ras-Raf-MEK-ERK cascade is a complex pathway – not only because it is swamped with acronyms. One term is often used to designate multiple isoforms; Ras and Raf each have three isoforms, MEK can exist as MEK1 or MEK2, while ERK refers to ERK1 and ERK2.4,5,6 The regulation of the Ras signaling pathway is also complex; in addition to EGF, there are other receptor tyrosine kinases (RTKs) capable of influencing the pathway. “With feedback existing at multiple levels, cells can bypass or select for additional mutations to acquire resistance,” says Marie Parsons, postdoctoral associate in medicine, Weill Cornell Medicine. Downstream, ERK regulates more than 250 substrates including transcription factors, protein kinases and phosphatases, cytoskeletal elements and apoptosis regulators.7
How does Ras signaling become dysregulated and lead to cancer?
When the Ras switch is permanently turned “on”, the resulting constitutive signaling provides the proliferative signals that enable tumorigenesis. This constitutive state can result from mutations in genes coding for key proteins in the pathway, including Ras itself. “Deregulation of the network most commonly occurs through activating mutations in Ras or the RTK-Ras signaling pathway” says Marina Pajic, associate professor and laboratory head, Personalised Cancer Therapeutics Laboratory, Garvan Institute of Medical Research. Pajic refers to the cBioPortal for Cancer Genomics and The Cancer Genome Atlas (TCGA) – both of which are home to large-scale cancer genomics datasets and enable patterns to be identified and quantified. For example, one TCGA-based study demonstrated the RTK-Ras pathway has the highest median frequency of mutations across all cancer types, with mutations found in up to 46% of human cancers.8
RAS genes are among the most frequently mutated genes in human cancers, and are typically associated with a single mutation at codons 12, 13 or 61, all of which favor the formation of active (GTP-bound) Ras.9,10 Despite having similar functional consequences, not all Ras mutants are created equal and occur at different frequencies: K-Ras is the predominantly mutated isoform (85%), followed by N-Ras (11%).11 “In certain cancers, like pancreatic ductal adenocarcinoma (PDAC), mutationally activated K-Ras is the most frequently occurring alteration. As we12,13,14 and others15 have shown, mutationally activated KRAS is found in 94% of cases and is a major contributor to therapeutic resistance,” says Pajic. Meanwhile, NRAS mutations predominate in melanoma.16,17 As RAS mutations are often tissue-specific, inhibitors that work well in one cancer type tend not to work in others, explains Parsons: “For example, EGFR inhibitors generally are more successful in melanoma than colorectal cancer which can be explained by other mutational factors.”
While RAS mutations are the most common, alterations can also occur at different or multiple stages of the pathway; approximately 60% of non-small cell lung cancer have alterations in “driver oncogenes” including EGFR, BRAF and KRAS.18 Meanwhile, 90% of pancreatic cancers are associated with KRAS mutations,19 and the majority of thyroid cancers are RAS or BRAF mutant.20 Overall, a constitutively active Ras-Raf-MEK-ERK pathway can lead to diverse outcomes, as the pathway controls genetic programs associated with cell growth, migration, survival and the cell cycle.21
Difficulties associated with targeting the Ras pathway
Currently, there are no available therapies specifically designed for Ras mutant cancers. The lack of clinical success, combined with the notorious difficulty associated with targeting the pathway, caused Ras to develop a reputation as being an “undruggable” target. While the “undruggable” title is now being challenged, key hurdles and complexities remain:22,23
- The topology of Ras is not suited to the design of high-affinity drugs, as it lacks an accessible pocket.
- Mutant proteins are almost identical to their essential, wild-type counterparts.
- Wild-type Ras isoforms can interact with oncogenic Ras proteins to display both tumor-promoting and tumor-suppressing functions.
“The pathway has been very tricky to target, particularly due to the cellular ability to rapidly overcome blockade, and reactivate signaling through downstream or parallel mechanisms,” reflects Pajic. Parsons adds: “While efforts have been made to develop small molecule inhibitors – some of which are in the clinic – they are often met with resistance, and cells persistently find a way to induce signaling even in the presence of an inhibitor. A lot of inhibitors are also met with variable responses in patients.”
Losing the “undruggable” label: Recent success and developments in Ras-targeted therapies
After a long bout of setbacks, recent success has generated renewed energy in the field. Promising Phase II results were observed where a KRASG12C inhibitor called sotorasib/AMG 510 led to rapid and durable tumor shrinkages in previously treated patients with advanced non-small cell lung cancer.24 “Now that we have seen it is possible,” says Parsons, “I think many efforts to improve this inhibitor will be underway. The 12C inhibitor isn’t perfect and papers already show resistance, but I think the fundamental concepts will open doors to the development of improved inhibitors with increased efficacy. Companies will keep this information under wraps for now, but I think many will build upon the initial findings.” The small molecule drug was the first KRASG12C inhibitor to enter the clinic and has been shown to inhibit KRAS signaling and tumor growth in cell lines and patient-derived xenograft models.25
As cells find new ways to develop resistance, researchers look to new tools and approaches – often beyond just a single solution. For instance, the use of MEK inhibitors alongside BRAF inhibitors in melanoma has been shown to slow the acquisition of resistance.26 Pajic points to a number of other developments, including the production of clinical grade “iExosomes” (mesenchymal stromal cell-derived exosomes with KrasG12D siRNA) for PDAC, and the targeting of KRAS effectors such as PTPN11 with first-in-human studies underway in patients with advanced solid cancers (e.g., NCT04528836, NCT04252339).27 Parson and colleagues are also exploring combination therapies across a range of preclinical models, and described how a KRAS12C inhibitor and EGFR inhibitor resulted in cell death in vitro.28
Some novel approaches to Ras-targeted therapies have involved the revival of compounds that were initially deemed unsuccessful. The development of Raf-MEK inhibitor CH5126766 looked promising at first, but was thwarted by toxicity issues. To explore potential ways of limiting toxicity, the inhibitor was profiled in a Phase I study using intermittent dosing schedules.29 Overall, it was concluded that antitumor activity was achieved across multiple cancer types, and the inhibitor was deemed tolerable – warranting further evaluation of the Raf-MEK inhibitor.
Persistent, creative thinking is what the field needs, which is what Pajic’s group is working to deliver. By exploring the use of novel, small molecule, stroma-targeting agents Pajic hopes to deprive cancer cells of the supportive stromal niche in primary tumors and metastases, while supporting chemotherapy and/or immuno-oncology agents.30,31
Precise, novel oncology tools and preclinical models are needed to drive the development of Ras-related therapeutics. For Parsons, her mission is to develop more representative animal models for colorectal cancer, as many current models develop tumors in the small intestine. Meanwhile, colleague and co-author Maria Zafra is using base editing to make distinct alterations in mice with specific KRAS mutations – now published in Cancer Discovery:28 “The mice and subsequent organoid cultures are a really valuable tool that faithfully recapitulate signaling dynamics, and serve as an important preclinical model. Previously, most studies would use cell lines which removes a lot of the complexities of signaling,” says Parsons. In the same laboratory, a reporter was developed to indicate when precise, inducible base editing has occurred. “This would be really exciting as you could introduce a specific mutation and then “turn off” the delivery system to avoid long term or off-target effects,” explains Parsons. The work opens the doors to implementing CRISPR base editing to create targeted mutations in cell lines, organoids and animal models.32
From the KRASG12C inhibitor breakthrough to the exploration of combination therapies, there has been significant progress in Ras-related research. Together, preclinical research tools such as base editing reporters, organoids and precise animal models are paving the way for the development of rational, tailored anticancer strategies.
“Apart from that I think the gene engineering field in general is driving this field forward,” says Parsons. “The way preclinical models are being developed is rapidly changing to make more representative models. The way we can induce mutations in organoids is changing and the CRISPR/base editing field is evolving at such a rapid pace. Many companies are also getting smarter with drug design and building upon existing drugs like the 12C inhibitor.”
References
1. Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020;80(14):2969-2974. doi: 10.1158/0008-5472.CAN-19-3682
2. Ghobrial IM, Adjei AA. Inhibitors of the Ras oncogene as therapeutic targets. Hematol Oncol Clin North Am. 2002;16(5):1065-1088. doi: 10.1016/S0889-8588(02)00050-3
3. Chang F, Steelman LS, Lee JT, et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17(7):1263-1293. doi: 10.1038/sj.leu.2402945
4. Rebocho AP, Marais R. ARAF acts as a scaffold to stabilize BRAF:CRAF heterodimers. Oncogene. 2013;32(26):3207-3212. doi: 10.1038/onc.2012.330
5. Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15(10):577-592. doi: 10.1038/nrc4000
6. Buscà R, Pouysségur J, Lenormand P. ERK1 and ERK2 MAP kinases: specific roles or functional redundancy? Front Cell Dev Biol. 2016;4. doi: 10.3389/fcell.2016.00053
7. Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24(1):21-44. doi: 10.1080/02699050500284218
8. Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321-337.e10. doi: 10.1016/j.cell.2018.03.035
9. Scott AJ, Lieu CH, Messersmith WA. Therapeutic approaches to RAS mutation. The Cancer Journal. 2016;22(3):165-174. doi: 10.1097/PPO.0000000000000187
10. Quinlan MP, Settleman J. Isoform-specific Ras functions in development and cancer. Future Oncol. 2009;5(1):105-116. doi: 10.2217/14796694.5.1.105
11. Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129(7):1287-1292. doi: 10.1242/jcs.182873
12. Bailey P, Chang DK, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47-52. doi: 10.1038/nature16965
13. Waddell N, Pajic M, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495-501. doi: 10.1038/nature14169
14. Biankin AV, Waddell N, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399-405. doi: 10.1038/nature11547
15. Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801-1806. doi: 10.1126/science.1164368
16. Stalnecker CA, Der CJ. RAS, wanted dead or alive: advances in targeting RAS mutant cancers. Sci Signal. 2020;13(624):eaay6013. doi: 10.1126/scisignal.aay6013
17. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13(11):828-851. doi: 10.1038/nrd4389
18. Zaman, Wu, Bivona. Targeting oncogenic BRAF: past, present, and future. Cancers. 2019;11(8):1197. doi: 10.3390/cancers11081197
19. Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059
20. The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676-690. doi: 10.1016/j.cell.2014.09.050
21. Gimple RC, Wang X. RAS: Striking at the core of the oncogenic circuitry. Front Oncol. 2019;9:965. doi: 10.3389/fonc.2019.00965
22. McCormick F. Targeting KRAS directly. Annu Rev Cancer Biol. 2018;8:81-90. doi: 10.1146/annurev-cancerbio-050216-122010
23. Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol. 2016;58:60-69. doi: 10.1016/j.semcdb.2016.07.012
24. Hong DS, Fakih MG, Strickler JH, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207-1217. doi: 10.1056/NEJMoa1917239
25. Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575(7781):217-223. doi: 10.1038/s41586-019-1694-1
26. Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8(1):48-56. doi: 10.1177/1758834015616934
27. Ruess DA, Heynen GJ, Ciecielski KJ, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24(7):954-960. doi: 10.1038/s41591-018-0024-8
28. Zafra MP, Parsons MJ, Kim J, et al. An in vivo Kras allelic series reveals distinct phenotypes of common oncogenic variants. Cancer Discov. 2020;10(11):1654-1671. doi: 10.1158/2159-8290.CD-20-0442
29. Guo C, Chénard-Poirier M, Roda D, et al. Intermittent schedules of the oral RAF–MEK inhibitor CH5126766/VS-6766 in patients with RAS/RAF-mutant solid tumours and multiple myeloma: a single-centre, open-label, phase 1 dose-escalation and basket dose-expansion study. Lancet Oncol. 2020;21(11):1478-1488. doi: 10.1016/S1470-2045(20)30464-2
30. Parkin A, Man J, Timpson P, Pajic M. Targeting the complexity of Src signalling in the tumour microenvironment of pancreatic cancer: from mechanism to therapy. FEBS J. 2019;286(18):3510-3539. doi: 10.1111/febs.15011
31. Vennin C, Murphy KJ, Morton JP, Cox TR, Pajic M, Timpson P. Reshaping the tumor stroma for treatment of pancreatic cancer. Gastroenterology. 2018;154(4):820-838. doi: 10.1053/j.gastro.2017.11.280
32. Katti A, Foronda M, Zimmerman J, et al. GO: a functional reporter system to identify and enrich base editing activity. Nucleic Acids Res. 2020;48(6):2841-2852. doi: 10.1093/nar/gkaa124


