Nanodiscs: The Mobile Home for Membrane Proteins

Complete the form below to unlock access to ALL audio articles.
The problem
Membrane proteins are a highly relevant research topic in the field of medicine. Most drug studies are based on identifying how a drug can influence the membrane protein in question. As the cells' connection to the extracellular world, membrane proteins sit at the top of numerous essential signaling chains involved in the cells physiological function.1 Thus, it is essential to study the membrane proteins both from a structural and functional perspective. However, this is much more difficult than working with their cytosolic counterparts.
One such major difficulty is keeping membrane proteins stable outside the cell membrane. This issue dates back to the one of the first structural studies of a membrane protein in 1985 by Deisenhofer et al.2 Since then, membrane proteins have remained a challenge for researchers in structural biology and other disciplines.
The main problem lies in the overall structure of membrane proteins. In contrast to soluble proteins, membrane proteins have hydrophobic amine acid residues on their surfaces, instead of polar ones. This enables better interaction with the surrounding hydrophobic chains of the phospholipids inside the membrane bilayer. If these hydrophobic amino acids residues should ever be surrounded by polar media, the membrane protein becomes instable and loses functionality. Such polar media includes water and almost every buffer that is involved in traditional protein purification. Therefore, the hydrophobic side chains of the membrane protein surface must be shielded from the hydrophilic surroundings.
Traditionally in the field of membrane protein research, this was achieved through the use of many different amphipathic detergents, such as Octylthioglucoside (OTG) or LDAO (Dodecyldimethylaminocid), that the membrane protein of interest (MPOI) was surrounded with and kept stable.
Figure 1 show a schematic depiction of this process. The disadvantages of this outdated procedure are numerous. First and foremost, not every protein reacts as well as other proteins to certain detergents. Usually a long and tedious screening process lies at the beginning of most detergent-based membrane protein assays. Furthermore, even if the membrane protein is stable surrounded by the detergent, the detergent might interfere with the functionality of the membrane protein and subsequently falsify any gathered data. Additionally, new detergent has to be constantly added to the proteins, otherwise they would destabilize. Seddon et al's 2004 review comprehensively summarizes these problems.

Figure 1: Membrane proteins (orange), whose hydrophobic side chains are covered by amphipathic detergents (black).
The solution: Nanodiscs
The solution to these problems was first described by Bayburt and colleagues in 1998: A small disc-shaped phospholipid bilayer system that mimics the cell membrane, known as “nanodiscs”.4 Today, scientists distinguish between two types of nanodiscs: Membrane-scaffolding-protein nanodiscs (MSP Nanodiscs) and synthetic nanodiscs.
A nanodisc is mainly defined by its components that hold the phospholipids together. In the case of the MSP nanodisc the phospholipid bilayer is held together by so called MSP (see Figure 2). These proteins derive from theapolipoprotein (apo) A-1 that is normally involved in the transport of cholesterol in the body. Depending on the MSP that has been used, the diameter of these nanodiscs can vary from seven to up to 50 nanometers.
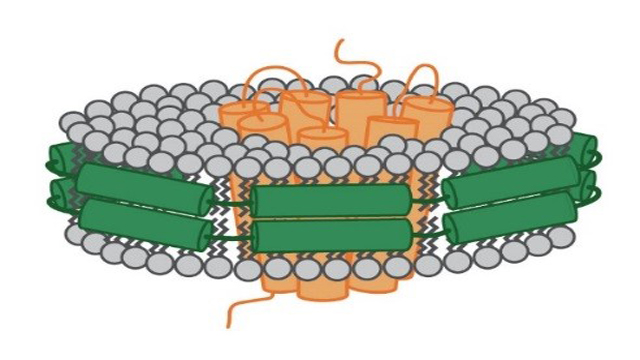
Figure 2: Schematic depiction of a MSP nanodisc. The membrane protein (orange) sits in the middle of the disc. It is surrounded by a ring made of phospholipids (grey). This ring is held together by membrane scaffolding proteins (green).
As this bilayer is composed of phospholipids, a nanodisc mimics the cell membrane in its composition. Therefore, it provides an environment representative of the native cell membrane better than any detergent could and prevents the described loss of functionality that could happen when working with detergents.
There are three ways to stabilize a membrane protein with an MSP nanodisc, as presented in Figure 3.
The first way is to avoid the cell membrane to begin with, using a pre-assembled nanodisc that automatically takes up the membrane protein. Expressing the MPOI in a cell-free method ensures that it is never located inside a cell membrane at all.
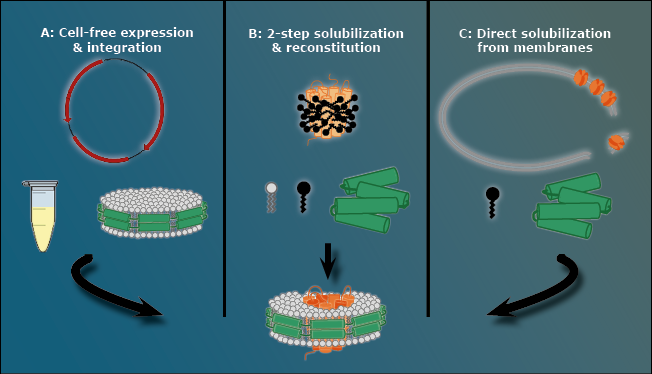 Figure 3: The three methods to create MSP nanodiscs. A: The cell-free approach. Using a pre-assembled nanodisc cell free expressed membrane proteins directly integrate into the cell membrane. B: Two-step solubilization and reconstitution. In this case a membrane protein that is already covered in detergent (black) is integrated into an MSP nanodisc that forms out of loose MSPs (green) and free phospholipids. C: Direct solubilization from cell membrane. In this case detergents (black) dissolve the cell membrane that contains the MPOI and during this process a loose MSP (green) forms a nanodisc out of the cell membrane’s own phospholipids with the MPOI inside.
Figure 3: The three methods to create MSP nanodiscs. A: The cell-free approach. Using a pre-assembled nanodisc cell free expressed membrane proteins directly integrate into the cell membrane. B: Two-step solubilization and reconstitution. In this case a membrane protein that is already covered in detergent (black) is integrated into an MSP nanodisc that forms out of loose MSPs (green) and free phospholipids. C: Direct solubilization from cell membrane. In this case detergents (black) dissolve the cell membrane that contains the MPOI and during this process a loose MSP (green) forms a nanodisc out of the cell membrane’s own phospholipids with the MPOI inside.
The second way is a two-step solubilization followed by a reconstitution step. In this procedure, the membrane protein is covered by detergent and all necessary components of a nanodisc (MSP and phospholipid) are added to the mixture. The MSP subsequently forms a nanodisc using the loose phospholipids. The detergent slowly falls of the membrane protein naturally and, after a given time, the membrane protein slips into the MSP nanodisc, similar to the cell free approach.
The third way is a direct solubilization from the cell membrane. Detergents are used to break up the cell membrane with an MSP present in the mixture. However, before the detergent removes all lipids around the cell membrane, the MSP forms the nanodiscs using the native cell membrane lipids. This option is the method with the closest native environment for the MPOI when it comes to MSP nanodiscs.
Synthetic nanodiscs
Synthethic nanodiscs serve the same purpose as MSP nanodiscs, however their creation method is different. Synthetic nanodiscs are based on SMALPS (styrene maleic acid lipid particles). One particular SMALP has been proven to be especially suited for synthetic nanodisc production. It is a synthetic Co-Polymere called Diisobutylene Maleic Acid (DIBMA).
DIBMA works functionally in the same way as MSP proteins in MSP nanodiscs. It holds the phospholipid ring together, as illustrated in Figure 4. DIBMA creates the synthetic nanodiscs in a format similar to how MSP nanodiscs are created (as depicted in figure 3C), but it functions as both the detergent and scaffolding protein. To use a metaphor, DIBMA works as a cookie cutter than cuts the MPOI out of the cookie dough (cell membrane), but stays around its piece to stabilize it.
 Figure 4: Synthetic nanodisc, based on DIBMA (blue). DIBMA is holding a bilayer circle of phospholipids (grey) together that are surrounding and stabilizing a membrane protein (orange).
Figure 4: Synthetic nanodisc, based on DIBMA (blue). DIBMA is holding a bilayer circle of phospholipids (grey) together that are surrounding and stabilizing a membrane protein (orange).
This way, synthetic nanodiscs always use the native phospholipid environment of the MPOI and therefore always provides the best possible conditions for it.
Applications of nanodiscs
Both types of nanodiscs are vastly superior to the outdated method of detergents to stabilize membrane proteins after the cell membrane has been removed. Nanodiscs can either use artificial lipid compositions or the native phospholipid environment of the membrane protein of interest. This ensures that even removed from the cell membrane, membrane protein remain structurally and functionally intact.
Nanodiscs have been used for multiple purposes. For example, in electron microscopy studies to analyze the light harvesting complex II (LHCII) in spinach, and in cancer research.5,6 Analyzing LHCII in nanodiscs brought new insights regarding its conformational flexibility that were previously unobservable in detergent based studies. In cancer research, nanodisc showed to be a promising and superior delivery tool to get the chemotherapeutic reagent DOX past the immune checkpoint of cancer cells.
References
1. Yin & Flynn. Drugging Membrane Protein Interactions. Annual Review of Biomedical Engineering. (2016) 18. 10.1146/annurev-bioeng-092115-025322.
2. Deisenhofer et al. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature (1985) 10.1038/318618a0
3. Seddon, A. et al. Membrane Proteins, Lipids and Detergents: Not Just a Soap Opera. Biochimica et biophysica acta. (2004) 1666. 105-17. 10.1016/j.bbamem.2004.04.011.
4. Bayburt, T.H. et al. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J. Struct. Biol. (1998), 123(1):37-44
5. Kuai, R. et al. Elimination of established tumors with nanodisc-based combination chemoimmunotherapy. Science Advances. 4. (2018). eaao1736. 10.1126/sciadv.aao1736.
6. Pandit A., et al. Assembly of the major light-harvesting complex II in lipid nanodiscs. Biophys. J. (2011) 101:2507-2515

