Cold Spring Harbor, NY — Scientists at Cold Spring Harbor Laboratory (CSHL) have discovered a way to rein in an overactive protein that drives some aggressive leukemias. The renegade molecule, MEF2C, belongs to a class of proteins that is notoriously difficult to manipulate with drugs. But the new research suggests an opportunity to develop therapies against it.
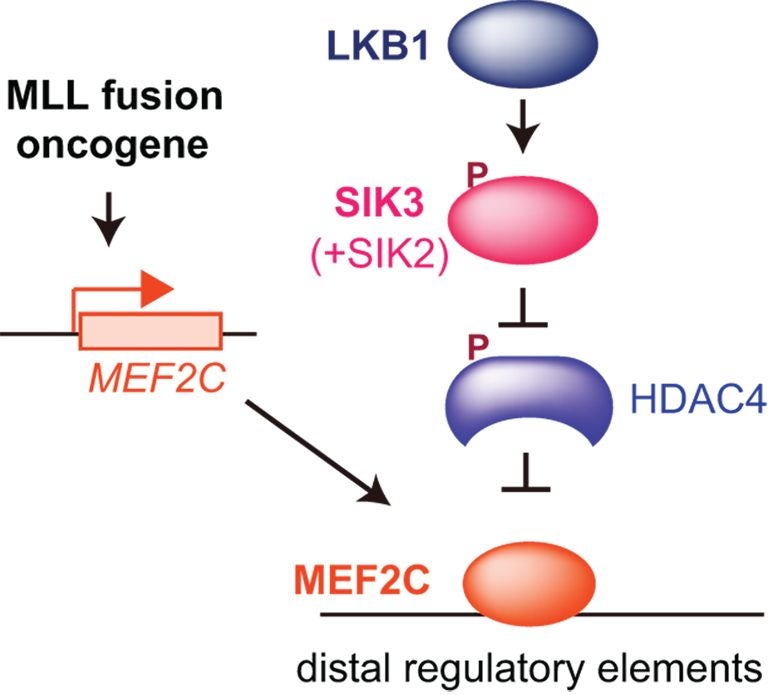
CSHL Associate Professor Christopher Vakoc, M.D., Ph.D., and colleagues now report that they can stop the growth of MEF2C-driven AML cells by blocking either of two target enzymes, known as LKB1 and salt-inducible kinase. The chemicals that the team used to interfere with the enzymes are small molecules that have the potential to be developed into drugs, Vakoc says.
The discovery is the result of a broad search for potential therapeutic strategies against AML that began several years ago in Vakoc’s lab. In 2013, his team devised a system based on CRISPR gene editing tools that they used to screen large numbers of genes, seeking to discover their impact on cancer cell survival. “We just let the cancer cells tell us what types of genes they need in order to grow,” Vakoc says.
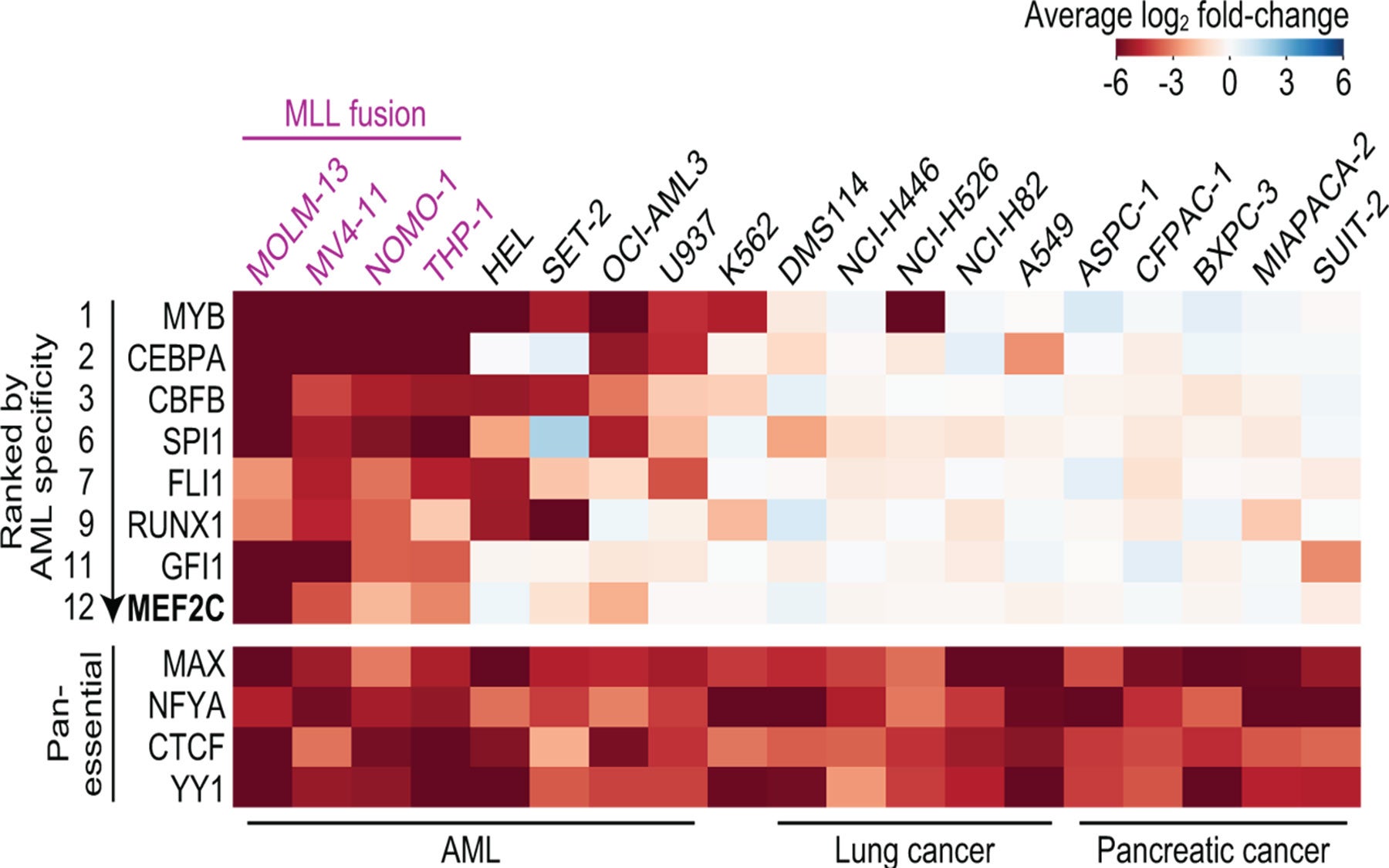
Led by Yusuke Tarumoto, a postdoctoral researcher in Vakoc’s lab, the team has now deployed that technology against AML. Their screens revealed that LKB1 and salt-inducible kinase are critical for the survival of certain AML cells. The enzymes had not previously been linked to AML, but with further experiments, the team learned that both help control the MEF2C transcription factor, which is a known cancer promoter.
“At the end of project, we realized we’d had actually discovered a way to control a transcription factor,” says Vakoc. That’s exciting, he says, because while most leukemias are thought to be caused by wayward transcription factors, such proteins are among the most challenging to target with drugs.
The chemicals the team used to switch off LKB1 and salt-inducible kinase in their lab-grown cancer cells are not suitable as therapeutic compounds, but Vakoc is optimistic that it will be possible to develop drugs that target these enzymes. Animal experiments are already underway in his lab to begin to investigate the strategy as a potential treatment approach.
Written by: Jennifer Michalowski, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
Cold Spring Harbor Laboratory NCI Cancer Center Support grant; Forbeck Foundation; Pershing Square Sohn Cancer Research Alliance; National Institutes of Health; National Cancer Institute; Leukemia & Lymphoma Society
Citation
Tarumoto Y et al, “LKB1, Salt-Inducible Kinases, and MEF2C are linked dependencies in acute myeloid leukemia,” appears online March 8, 2018 in Molecular Cell.
Principal Investigator

Chris Vakoc
Professor
Alan and Edith Seligson Professor of Cancer Research
Cancer Center Deputy Director of Research
M.D., Ph.D., University of Pennsylvania, 2007