An Introduction to PCR

Complete the form below to unlock access to ALL audio articles.
Contents
Standard PCR experiment overview
- Quantitative real-time PCR (qPCR)
- Reverse transcription-PCR (RT-PCR)
- Reverse transcription-quantitative PCR (RT-qPCR)
- Digital PCR (dPCR) and digital droplet PCR (ddPCR)
- Microfluidic PCR
What is PCR?
Polymerase chain reaction (PCR) is a technique that has revolutionized the world of molecular biology and beyond, enabling the amplification of nucleotide sequences. In this article, we will discuss a brief history of PCR and its principles, highlighting the different types of PCR and the specific purposes to which they are being applied.
PCR principles and history
In 1983, American biochemist Kary Mullis was driving home late at night when a flash of inspiration struck him. He wrote on the back of a receipt the idea that would eventually grant him the Nobel Prize for Chemistry in 1993. The concept was straightforward: reproducing in a laboratory tube the DNA replication process that takes place in cells. The outcome is the same: the generation of new complementary DNA (cDNA) strands based upon the existing ones.
Mullis used the basis of Sanger's DNA sequencing as a starting point for his new technique. He realized that the repeated use of DNA polymerase triggered a chain reaction resulting in a specific DNA segment's amplification.
The foundations for his idea were laid by a discovery in 1976 of a thermostable DNA polymerase, Taq, isolated from the bacterium Thermus aquaticus found in hot springs.1 Taq DNA polymerase has a temperature optimum of 72 °C and survives prolonged exposure to temperatures as high as 96 °C, meaning that it can tolerate several denaturation cycles.
Before the discovery of Taq polymerase, molecular biologists were already trying to optimize cyclic DNA amplification protocols, but they needed to add fresh polymerase at each cycle because the enzyme could not withstand the high temperatures needed for DNA denaturation. Having a thermostable enzyme meant that they could repeat the amplification process many times over without the need for fresh polymerase at every cycle, making the whole process scalable, more efficient and less time-consuming.
The first description of this polymerase chain reaction (PCR) using Taq polymerase was published in Science in 1985.2
In 1993, the first FDA-approved PCR kit came to market. Since then, PCR has been steadily and systematically improved. It has become a game-changer in everything from forensic evidence analysis and diagnostics, to disease monitoring and genetic engineering. It is undoubtedly considered one of the most important scientific advances of the 20th century.
Standard PCR experiment overview
The PCR is used to amplify a specific DNA fragment from a complex mixture of starting material called template DNA. The sample preparation and purification protocols depend on the starting material, including the sample matrix and accessibility of target DNA. Often, minimal DNA purification is needed and some techniques such as direct PCR or extraction-free PCR require no pre-purification of DNA or RNA. However, PCR does require knowledge of the DNA sequence information that flanks the DNA fragment to be amplified (called target DNA).
From a practical point of view, a PCR experiment is relatively straightforward and can be completed in a few hours. In general, a PCR reaction needs five key reagents:
DNA to be amplified: also called PCR template or template DNA. This DNA can be of any source, such as genomic DNA (gDNA), cDNA, and plasmid DNA.
DNA polymerase: all PCR reactions require a DNA polymerase that can work at high temperatures. Taq polymerase is a commonly used one, which can incorporate nucleotides at a rate of 60 bases/second at 70 °C and can amplify templates of up to 5 kb, making it suitable for standard PCR without special requirements. New generations of polymerases are being engineered to improve reaction performance. For example, some are engineered to be only activated at high temperatures to reduce non-specific amplification at the beginning of the reaction. Others incorporate a “proofreading” function, important, for example, when it is critical that the amplified sequence matches the template sequence exactly, such as during cloning.
Primers: DNA polymerases require a short sequence of nucleotides to indicate where they need to initiate amplification. In a PCR, these sequences are called primers and are short pieces of single-stranded DNA (approximately 15-30 bases). When designing a PCR experiment, the researcher determines the region of DNA to be amplified and designs a pair of primers, one on the forward strand and one on the reverse, that specifically flanks the target region. Primer design is a key component of a PCR experiment and should be done carefully. Primer sequences must be chosen to target the unique DNA of interest, avoiding the possibility of binding to a similar sequence. They should have similar melting temperatures because the annealing step occurs simultaneously for both strands. The melting temperature of a primer can be impacted by the percentage of bases that are guanine (G) or cytosine (C) compared to adenine (A) or thymine (T), with higher GC contents increasing melting temperatures. Adjusting primer lengths can help to compensate for this in matching a primer pair. It is also important to avoid sequences that will tend to form secondary structures or primer dimers, as this will reduce PCR efficiency. Many free online tools are available to aid in primer design.
Deoxynucleotide triphosphates (dNTPs): these serve as the building blocks to synthesize the new strands of DNA and include the four basic DNA nucleotides (dATP, dCTP, dGTP, and dTTP). dNTPs are usually added to the PCR reaction in equimolar amounts for optimal base incorporation.
PCR buffer: the PCR buffer ensures that optimal conditions are maintained throughout the PCR reaction. The major components of PCR buffers include magnesium chloride (MgCl2), tris-HCl and potassium chloride (KCl). MgCl2 serves as a cofactor for the DNA polymerase, while tris-HCl and KCl maintain a stable pH during the reaction.
The PCR reaction is carried out in a single tube by mixing the reagents mentioned above and placing the tube in a thermal cycler.
The PCR amplification consists of three defined sets of times and temperatures termed steps: denaturation, annealing, and extension (Figure 1).
 Figure 1: Steps of a single PCR cycle.
Figure 1: Steps of a single PCR cycle.
Each of these steps, termed cycles, is repeated 30-40 times, , doubling the amount of DNA at each cycle and obtaining amplification (Figure 2).

Figure 2: The different stages and cycles of DNA molecule amplification by PCR.
Let's take a closer look at each step.
1. Denaturation
The first step of PCR, called denaturation, heats the template DNA up to 95 °C for a few seconds, separating the two DNA strands as the hydrogen bonds between them are rapidly broken.
2. Annealing
The reaction mixture is then cooled for 30 seconds to 1 minute. Annealing temperatures are usually 50 - 65 °C however, the exact optimal temperature depends on the primers' length and sequence. It must be carefully optimized with every new set of primers.
The two DNA strands could rejoin at this temperature, but most do not because the mixture contains a large excess of primers that bind, or anneal, to the template DNA at specific, complementary positions. Once the annealing step is completed, hydrogen bonds will form between the template DNA and the primers. At this point, the polymerase is ready to extend the DNA sequence.
3. Extension
The temperature is then raised to the ideal working temperature for the DNA polymerase present in the mixture, typically around 72 °C, 74 °C in the case of Taq.
The DNA polymerase attaches to one end of each primer and synthesizes new strands of DNA, complementary to the template DNA. Now we have four strands of DNA instead of the two that were present to start with.
The temperature is raised back to 94 °C and the double-stranded DNA molecules – both the "original" molecules and the newly synthesized ones – denature again into single strands. This begins the second cycle of denaturation-annealing-extension. At the end of this second cycle, there are eight molecules of single-stranded DNA. By repeating the cycle 30 times, the double-stranded DNA molecules present at the beginning are converted into over 130 million new double-stranded molecules, each one a copy of the region of the starting molecule delineated by the annealing sites of the two primers.
To determine if amplification has been successful, PCR products may be visualized using gel electrophoresis, indicating amplicon presence/absence, size and approximate abundance. Depending on the application and the research question, this may be the endpoint of an experiment, for example, if determining whether a gene is present or not. Otherwise, the PCR product may just be the starting point for more complex downstream investigations such as sequencing and cloning.
PCR variations
Thanks to their versatility, PCR techniques have evolved over recent years leading to the development of several different types of PCR technology.
Some of the most widely used ones are:
Quantitative real-time PCR (qPCR)
One of the most useful developments has been quantitative real-time PCR or qPCR. As the name suggests, qPCR is a quantitative technique that allows real-time monitoring of the amplification process and detection of PCR products as they are made.2 It can be used to determine the starting concentration of the target DNA, negating the need for gel electrophoresis in many cases. This is achieved thanks to the inclusion of non-specific fluorescent intercalating dyes, such as SYBR® Green, that fluoresce when bound to double-stranded DNA, or DNA oligonucleotide sequence-specific fluorescent probes, such as hydrolysis (TaqMan) probes and molecular beacons. Probes bind specifically to DNA target sequences within the amplicon and use the principle of Förster Resonance Energy Transfer (FRET) to generate fluorescence via the coupling of a fluorescent molecule on one end and a quencher at the other end. For both fluorescent dyes and probes, as the number of copies of the target DNA increases, the fluorescence level increases proportionally, allowing real-time quantification of the amplification with reference to standards containing known copy numbers (Figure 3).
qPCR uses specialized thermal cyclers equipped with fluorescent detection systems that monitor the fluorescent signal as the amplification occurs.
Figure 3: Example qPCR amplification plot and standard curve used to enable quantification of copy number in unknown samples.
Reverse transcription-PCR (RT-PCR)
Reverse transcription (RT) -PCR and RT-qPCR are two commonly used PCR variants enabling gene transcription analysis and quantification of viral RNA, both in clinical and research settings.
RT is the process of making cDNA from single-stranded template RNA3 and is consequently also called first-strand cDNA synthesis. The first step of RT-PCR is to synthesize a DNA/RNA hybrid between the RNA template and a DNA oligonucleotide primer. The reverse transcriptase enzyme that catalyzes this reaction has RNase activity that then degrades the RNA portion of the hybrid. Subsequently, a single-stranded DNA molecule is synthesized by the DNA polymerase activity of the reverse transcriptase. High purity and quality starting RNA are essential for a successful RT-PCR.
RT-PCR can be performed following two approaches: one-step RT-PCR and two-step RT-PCR. In the first case, the RT reaction and the PCR reaction occur in the same tube, while in the two-step RT-PCR, the two reactions are separate and performed sequentially.
Reverse transcription-quantitative PCR (RT-qPCR)
The reverse transcription described above often serves as the first step in qPCR too, quantifying RNA in biological samples (either RNA transcripts or derived from viral RNA genomes).
As with RT-PCR, there are two approaches for quantifying RNA by RT-qPCR: one-step RT-qPCR and two-step RT-qPCR. In both cases, RNA is first reverse-transcribed into cDNA, which is used as the template for qPCR amplification. In the two-step method, the reverse transcription and the qPCR amplification occur sequentially as two separate experiments. In the one-step method, RT and qPCR are performed in the same tube.
Digital PCR (dPCR) and digital droplet PCR (ddPCR)
Digital PCR (dPCR) is another adaptation of the original PCR protocol.4 Like qPCR, dPCR technology uses DNA polymerase to amplify target DNA from a complex sample using a primer set and probes. The main difference, though, lies in the partitioning of the PCR reactions and data acquisition at the end.
dPCR and ddPCR are based on the concept of limiting dilutions. The PCR reaction is split into large numbers of nanoliter-sized sub-reactions (partitions). The PCR amplification is carried out within each droplet. Following PCR, each droplet is analyzed with Poisson statistics to determine the percentage of PCR-positive droplets in the original sample. Some partitions may contain one or more copies of the target, while others may contain no target sequences. Therefore, partitions classify either as positive (target detected) or negative (target not detected), providing the basis for a digital output format.
ddPCR is a recent technology that became available in 2011.5 ddPCR utilizes a water-oil emulsion to form the partitions that separate the template DNA molecules. The droplets essentially serve as individual test tubes in which the PCR reaction takes place. This technology was put to use in creating sensitive SARS-CoV-2 tests.
Microfluidic PCR
The recent development of microfluidic handling systems with microchannels and microchambers has paved the way for a range of practical applications, including the amplification of DNA via PCR on microfluidic chips.
PCR performed on a chip benefits from microfluidics’ advantages in speed, sensitivity and low consumption of reagents. These features make microfluidic PCR particularly appealing for point-of-care testing, for example, for diagnostics applications. From a practical point of view, the sample flows through a microfluidic channel, repeatedly passing the three temperature zones reflecting the different steps of PCR. It takes just 90 seconds for a 10 μL sample to perform 20 PCR cycles.6 The subsequent analysis can then be easily carried out off-chip.
PCR troubleshooting
The different PCR approaches all have advantages and disadvantages that impact the applications to which they are suited 7. These are summarized in Table 1.
| Approach | Advantages | Limitations |
| PCR | · Easiest PCR to perform · Low cost of equipment and reagents · Several downstream applications (e.g., cloning) | · Results are only qualitative · Requires post-amplification analyses that increase time and risk of error · Products may need to be confirmed by sequencing |
| qPCR | · Produces quantitative results · Probe use can ensure high specificity · High analytical sensitivity · Low turnaround time · Eliminates requirements for post-amplification analysis | · Requires more expensive reagents and equipment · Less flexibility in primer and probe selection · Less amenable to other downstream product confirmation analyses (such as sequencing) due to the small length of the amplicon · Not suitable for some downstream applications such as cloning
|
| RT-PCR and RT-qPCR | · Can be used with all RNA types | · RNA is prone to degradation · The RT step may increase the time and potential for contamination |
| dPCR and ddPCR | · Fast · No DNA purification step · Provides absolute quantification · Increased sensitivity for detecting the target in limited clinical samples · Highly scalable | · Costly · Based on several statistical assumptions |
| Microfluidic PCR | · Accelerated PCR process · Reduced reagent consumption · Can be adapted for high throughput · Portable device for point-of-care applications · Allows single-cell analysis | · Still very new technology · Requires extensive sample preparation to remove debris and unwanted compounds · Restricted choice of materials for the microfluidic device due to high temperatures |
PCR output applications
PCR has become an indispensable tool in modern molecular biology and has completely transformed scientific research. The technique has also opened up the investigation of cellular and molecular processes to those outside the field of molecular biology and consequently also finds utility by scientists in many disciplines.
Whilst PCR is itself a powerful standalone technique, it has also been incorporated into wider techniques, such as cloning and sequencing, as one small but important part of these workflows.
Research applications of PCR include:
Gene transcription - PCR can examine variations in gene transcription among cell types, tissues and organisms at a specific time point. In this process, RNA is isolated from samples of interest, and reverse-transcribed into cDNA. The original levels of RNA for a specific gene can then be quantified from the amount of cDNA amplified in PCR.
Genotyping - PCR can detect sequence variations in alleles of specific cells or organisms. A common example is the genotyping of transgenic organisms, such as knock-out and knock-in mice. In this application, primers are designed to amplify either a transgene portion (in a transgenic animal) or the mutation (in a mutant animal).
Cloning and mutagenesis - PCR cloning is a widely used technique where double-stranded DNA fragments amplified by PCR are inserted into vectors (e.g., gDNA, cDNA, plasmid DNA). This for example, enables the creation of bacterial strains from which genetic material has been deleted or inserted. Site-directed mutagenesis can also be used to introduce point mutations via cloning. This often employs a technique known as recombinant PCR, in which overlapping primers are specifically designed to incorporate base substitutions (Figure 4). This technique can also be used to create novel gene fusions.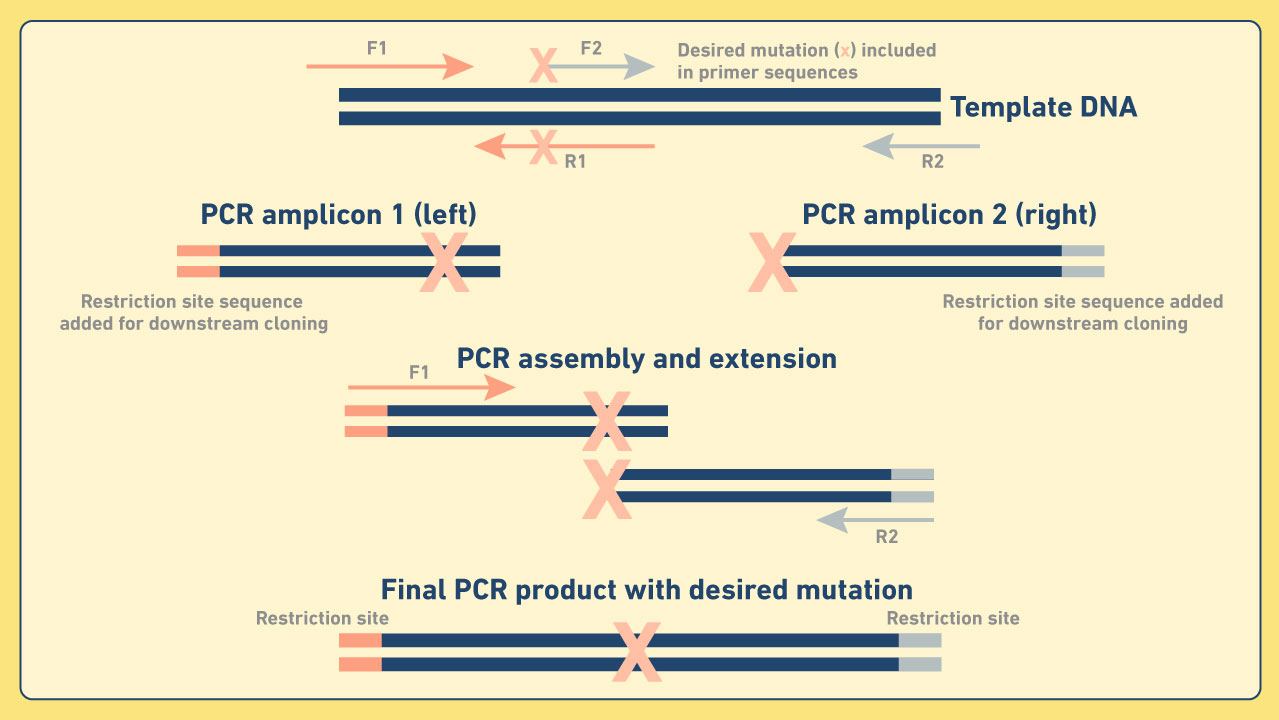 Figure 4: Diagram depicting an example of recombinant PCR.
Figure 4: Diagram depicting an example of recombinant PCR.
Sequencing - PCR can be used to enrich template DNA for sequencing. The type of PCR recommended for the preparation of sequencing templates is called high-fidelity PCR and is able to maintain DNA sequence accuracy. In Sanger sequencing, PCR-amplified fragments are then purified and run in a sequencing reaction. In next-generation sequencing (NGS), PCR is used at the library preparation stage, where DNA samples are enriched by PCR to increase the starting quantity and tagged with sequencing adaptors to allow multiplexing. Bridge PCR is also an important part of the second-generation NGS sequencing process.
Both as an independent technique and as a workhorse within other methods, PCR has transformed a range of disciplines. These include:
Genetic research - PCR is used in most laboratories worldwide. One of the most common applications is gene transcription analysis9, aimed at evaluating the presence or abundance of particular gene transcripts. It is a powerful technique in manipulating the genetic sequence of organisms – animal, plant and microbe - through cloning. This enables genes or sections of genes to be inserted, deleted or mutated to engineer in genetic markers alter phenotypes, elucidate gene functions and develop vaccines to name but a few. In genotyping, PCR can be used to detect sequence variations in alleles in specific cells or organisms. Its use isn’t restricted to humans either. Genotyping plants in agriculture assists plant breeders in selecting, refining, and improving their breeding stock. PCR is also the first step to enrich sequencing samples, as discussed above. For example, most mapping techniques in the Human Genome Project (HGP) relied on PCR.
Medicine and biomedical research - PCR is used in a host of medical applications, from diagnostic testing for disease-associated genetic mutations, to the identification of infectious agents. Another great example of PCR use in the medical realm is prenatal genetic testing. Prenatal genetic testing through PCR can identify chromosome abnormalities and genetic mutations in the fetus, giving parents-to-be important information about whether their baby has certain genetic disorders. PCR can also be used as a preimplantation genetic diagnosis tool to screen embryos for in vitro fertilization (IVF) procedures.
Forensic science - Our unique genetic fingerprints mean that PCR can be instrumental in both paternity testing and forensic investigations to pinpoint samples' sources. Small DNA samples isolated from a crime scene can be compared with a DNA database or with suspects' DNA, for example. These procedures have really changed the way police investigations are carried out. Authenticity testing also makes use of PCR genetic markers, for example, to determine the species from which meat is derived. Molecular archaeology too utilizes PCR to amplify DNA from archaeological remains.
Environmental microbiology and food safety - Detection of pathogens by PCR, not only in patients' samples but also in matrices like food or water, can be vital in diagnosing and preventing infectious disease.
PCR is the benchmark technology for detecting nucleic acids in every area, from biomedical research to forensic applications. Kary Mullis's idea, written on the back of a receipt on the side of the road, turned out to be a revolutionary one.
1. Chien A, Edgar DB, Trela JM. Deoxyribonucleic acid polymerase from the extreme
thermophile Thermus aquaticus. J Bacteriol. 1976;127(3):1550-57
doi: 10.1128/JB.127.3.1550-1557.1976
2. Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230(4732):1350 doi: 10.1126/science.2999980
3. Arya M, Shergill IS,
Williamson M, Gommersall L, Arya N, Patel HRH. Basic principles of real-time
quantitative PCR. Expert Rev Mol Diagn. 2005;5(2):209-19 doi: 10.1586/14737159.5.2.209
4. Bachman J. Chapter Two - Reverse-Transcription PCR (RT-PCR). In: Lorsch J,
ed. Methods in Enzymology: Academic Press, 2013:67-74. doi : 10.1016/B978-0-12-420037-1.00002-6
5. Morley AA. Digital PCR: A brief history. Biomol Detect Quantif.
2014;1(1):1-2 doi: 10.1016/j.bdq.2014.06.001
6. Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene
expression analysis with low abundant targets: from variable nonsense to
publication quality data. Sci Rep. 2017;7(1):2409 doi: 10.1038/s41598-017-02217-x
7. Ahrberg CD, Manz A, Chung BG. Polymerase chain reaction in microfluidic
devices. Lab Chip. 2016;16(20):3866-84 doi: 10.1039/C6LC00984K
8. Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol.
2013;133(3):1-4 doi: 10.1038/jid.2013.1
9. VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR
for gene expression analysis. BioTechniques. 2008;44(5):619-26 doi: 10.2144/000112776


