MALDI Imaging Empowers Omics in Spatial Context for Unprecedented Molecular Insight
Complete the form below to unlock access to ALL audio articles.
Analyzing the molecular profile of a single cell is essential for discovering mechanisms not seen when studying a bulk population of cells, particularly when that population is comprised of multiple cell types. As we know, single cells are anything but solitary, necessitating the study of how spatially structured communities of individual cells act and interact in the context of their networked environment.
Mass spectrometry (MS) techniques have become important analytical tools for proteomic and metabolomic analysis of single cells, and we have long recognized label-free matrix-assisted laser desorption/ionization (MALDI) MS imaging as a valuable tool for identifying and spatially correlating a variety of molecular compounds including metabolites, lipids and proteins directly from tissue. But the dynamic range of molecules within single cells present a challenge to its relatively low molecular sensitivity and specificity.
Proteomic analysis by liquid chromatography mass spectrometry (LC-MS) of subregions identified by MALDI imaging can provide high sensitivity omics studies but, until recently, such an approach has required two separate instruments. The spatial omics workflow, provides the opportunity to combine regiospecific information from MALDI imaging with deep proteomic coverage for biomarker discovery and molecular characterization.1
Spatial omics is now more efficient and economical because of recent advances in dual-source MS instrumentation that offer both MALDI and electrospray ionization (ESI) sources and enable next-level MALDI imaging and LC-MS capabilities on the same instrument.
The utility of novel spatial omics workflows using a single instrument is nicely demonstrated in a recent study looking at the microproteomic characterization of tumor subpopulations segmented by lipid profiles. Researchers at the University of Maastricht in the Netherlands have used this workflow to uncover proteomic profiles in tumor subpopulations of breast cancer.2
Heterogeneity in the tumor microenvironment can have implications both for tumor development and therapeutic outcomes. All too often, cells appear similar histologically but are fuelled by different biochemical engines that drive different tumorigenesis. We therefore need to be able to identify distinct tumor subpopulations using regiospecific molecular information. Spatial omics allows subtyping of tissue using thousands of label-free molecular images, and subsequent omics analysis helps us understand how individual cells act and interact in the context of their heterogeneous tumor environment.
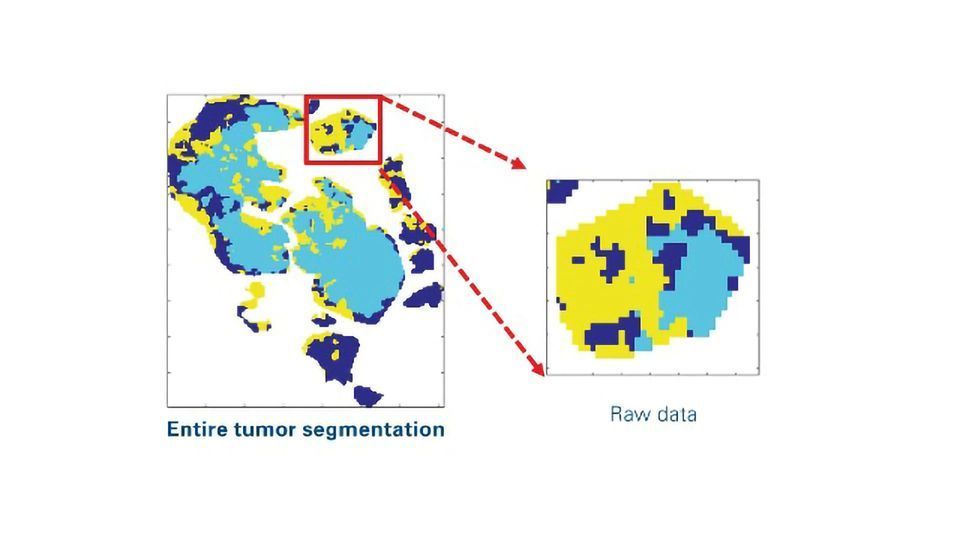
In this workflow, researchers used unsupervised segmentation of a panel of lipid images to subtype regions of interest (ROIs) in tissue sections (Figure 1). These image processing strategies provide boundary information of segments of tumor subpopulations for laser capture microdissection (LMD).
Figure 1: Image processing pipeline to produce ROI boundary information as x,y-coordinates for laser capture microdissection from segmentation raw data. The workflow includes smoothing, removal of small objects, holes filling, boundary detection and up-scaling to the optical image. Credit: Bruker Daltonics. Click image to view full size.
The study then proceeded with protein extraction and tryptic digestion of a small quantity of microdissected material (ca. 2,000 cells from each subregion) followed by proteomic analysis on a dual source trapped ion mobility (TIMS) quadrupole time-of-flight (QTOF) mass spectrometer using PASEF (parallel accumulation serial fragmentation). Here, PASEF allows extremely high-speed analysis without losing sensitivity or resolution – vitally important for analyzing very small microextracted regions. Spatial omics provides unprecedented insight to specific ROIs for microextraction and in-depth proteomics analysis to characterize regiospecific molecular changes across subtypes.
As well as spatial omics, advances in TIMS QTOF MS technology are enabling development of new MALDI imaging strategies. The recently introduced laser-based post-ionization (PI) technique, known as MALDI-2, provides a sensitivity boost that opens the door to imaging compounds typically inaccessible to traditional MALDI such as lipids, vitamins, and glycans.3 MALDI imaging is also popular for high-throughput analysis of oligosaccharides including N-linked glycans and, when MALDI-2 is implemented, sensitivity for compound images is increased by as much as three orders of magnitude.4
The combination of TIMS and MALDI-2 is uniquely powerful for spatial omics. Cellular content spans a diverse range of molecular classes, particularly when enhanced by MALDI-2, rendering TIMS separation essential to producing thousands of images from a single measurement. An added bonus is that the ion suppression which manifests in traditional MALDI imaging experiments is reduced with MALDI-2.
All these advantages are allowing researchers to look beyond the world of limited protein imaging to delve into the role that metabolites, lipids and even glycosylation patterns play in the development and pathogenesis of different cancers, including ovarian cancer, breast cancer, and myxoid liposarcoma.
The research summarized here includes a small portion of numerous examples of where the industry is heading. These techniques also have the potential to transform targeted drug and metabolite imaging in pharma studies, helping to make critical decisions in drug discovery, or to help bridge the gap between 4D-omics and pathology.
References
1. Dewez F, Oejten J, Henkel C, et al. Ms imaging‐guided microproteomics for spatial omics on a single instrument. Proteomics. 2020;20(23):1900369. doi:10.1002/pmic.201900369
2. Oetjen J, Hebeler R, Dewez F, Henkel C, Balluff B, Heeren R. MALDI guided SpatialOMx uncovers proteomic profiles in tumor subpopulations of breast cancer. [App Note]. Bruker Daltonics. 2020.
3. Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Muthing J, Dreisewerd K. Mass spectrometry imaging with laser-induced postionization. Science. 2015;348(6231):211-215. doi:10.1126/science.aaa1051
4. Heijs B, Potthoff A, Soltwisch J, Dreisewerd K. MALDI-2 for the enhanced analysis of N-linked glycans by mass spectrometry imaging. Anal Chem. 2020;92(20):13904-13911. doi:10.1021/acs.analchem.0c02732
About the author:
Shannon Cornett is Global MS Imaging Market Manager, Bruker Daltonics, Billerica, Massachusetts, USA.

