High-Throughput LC-MS for Early Drug Screening and Biomarker Detection

Complete the form below to unlock access to ALL audio articles.
High-throughput screening (HTS), or the rapid screening of large compound libraries for activity against biological targets, is a big part of the drug discovery process, especially in the early stages. There are millions of compounds with the potential to be a new drug and sifting through these compounds to identify a few with the highest potential – known as drug leads – could take months, if it were not for the current HTS techniques. These state-of-the-art techniques allow pharmaceutical researchers to accelerate the early drug screening process and reduce the time spent on discovering leads, so that more time and attention can be given to optimizing these leads for efficacy and safety.
Conducting large numbers of chemical tests expeditiously is not a strategy limited to drug discovery; researchers apply it in many different areas of biology and chemistry. Another example comes in the form of biomarker detection. In this article, we explore two novel high-throughput analytical methods based on liquid chromatography-mass spectrometry (LC-MS) analysis that can augment early drug screening and biomarker detection in the clinic.
LC-MS to rapidly screen drug binding
“The great thing about mass spectrometric screening methods is that they can detect compounds directly by mass, and so they are more reliable than other techniques that allow only indirect detection,” said Professor Heather Desaire, Department of Chemistry, University of Kansas, Kansas, USA. “For example, most spectroscopic techniques utilizing light to identify compounds have indirect detection schemes which sometimes interfere with results.”
Also, mass spectrometric methods are fast, selective, highly-sensitive (require only a small quantity of compounds), and can detect multiple compounds at a time. But the problem lies in the occurrence of false hits or misses, Desaire told us. “When these MS assays are used to identify potential lead compounds for target proteins, they suffer from a high rate of false positives and false negatives, which can lead to significant issues during later stages of the drug development cycle.”
In finding a way to tackle this, Desaire and her students decided to develop a new-and-improved mass spectrometric assay to probe for lead compounds. Their approach was focused on detecting a reporter compound that ionizes well, instead of detecting the lead itself, which may not ionize (1). This way, researchers don’t overlook any potential leads that are not ionizable, resulting in false negatives – one of the common problems in using mass spectrometry for HTS, according to Desaire and Dr. Kasun Imaduwage, lead author of the paper detailing the new method.
An interrogation technique to reduce false negatives
Desaire’s and Imaduwage’s new technique relies heavily on a ‘reporter’ molecule – a known, ionizable weak binder of the target that allows binding events of interest to be measured. The reporter is first incubated with the immobilized target protein. Once excess reporter is washed off, the reporter-target complex is introduced to a batch of library compounds. If the library contains a lead that is a stronger binder than the reporter, it competes for the same binding site in the protein and displaces the reporter from the complex. LC-MS is then used to detect and measure the displaced reporter molecules, after centrifugation.
When a high-affinity binder displaces a reporter, the reporter signal increases during detection, compared to control samples. (Figure 1) In this manner, a binding event is measurable, even if the high-affinity ligand is not detectable by MS. “This is an interrogation technique, where we are probing the presence of a strong binder in a library batch by detecting only the reporter molecule,” said Imaduwage.

Figure 1. The experimental workflow for Desaire’s and Imaduwage’s novel high-throughput LC-MS screening method. (Source: Imaduwage et al. Sci. Rep. 2017).
One challenge associated with this method is the identification of lower affinity ligands with equilibrium dissociation constants (Kd) in the micromolar range. It becomes especially difficult when the amount of target protein is limited. Another challenge is related to target immobilization. “For our assay, it’s very important to immobilize the target protein in order to separate unbound compounds from the protein-ligand complex,” explained Imaduwage. “However, immobilization requires several hours and confers numerous disadvantages. So, an automated experimental set-up that does not require target immobilization would be a promising approach to developing an ultra-high-throughput screening assay.” Desaire’s group believes that using advanced membrane filters may solve this issue.
“The most interesting outcome was identifying a new carbonic anhydrase (CA) inhibitor, pifthrin-µ,” remarked Imaduwage. “This is a significant result, considering that CA inhibitors are therapeutically valuable drugs for a number of diseases.” He went onto explain that pifthrin-µ can’t be detected by other HTS assays that employ electron spray ionization-mass spectrometry (ESI-MS), because it’s not ionizable. “That’s likely the reason that pifthrin- µ has remained under the radar, so to speak. It hasn’t been identified up until now as a CA inhibitor, even though other scientists have screened over 100,000 compounds in MS-based HTS assays, by using CA as a target protein.”
In closing, Desaire emphasized that time is not always the deciding factor for determining how good a method is for HTS, especially with the current availability of different sample preparation and separation techniques. “An effective HTS assay should be pertinent to different classes of chemical compounds. It should be faster because different compounds are assayed in the same run/batch, instead of in repeated runs.”
Therefore, choosing an assay that meets all these factors is vital, and as Desaire says, it is entirely dependent on the researcher to decide the proper technique based on the requirements of the desired experiment.
A high-throughput method to monitor alcohol misuse
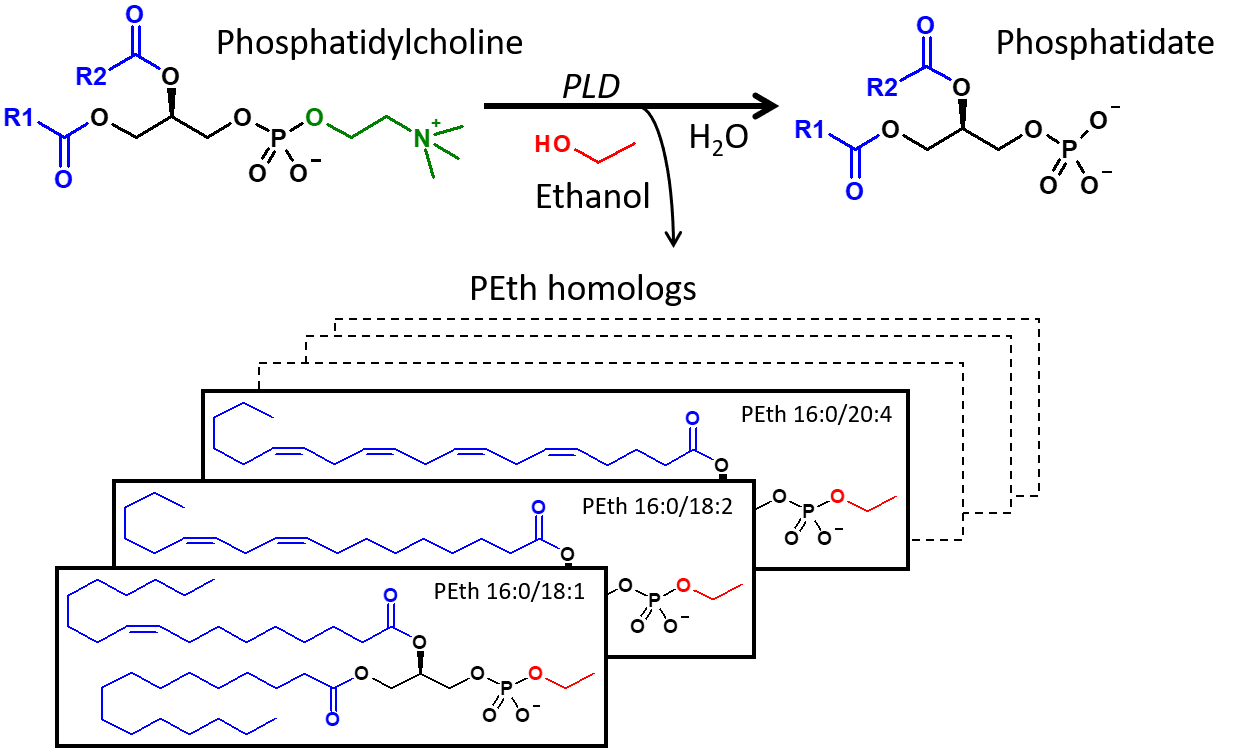
There are several different biomarkers that clinicians use to detect alcohol misuse in patients and phosphatidylethanol (PEth) is high on the list (2). Since its introduction to the clinic in 2006, PEth has been readily used to track alcohol consumption because of high clinical specificity and sensitivity, and a long window of detection. “PEth has higher clinical specificity and sensitivity than commonly used alcohol markers such as CDT or gamma-GT; the specificity is theoretically 100% since it is an ethanol metabolite,” said Associate Professor Anders Isaksson, Division of Clinical Chemistry and Pharmacology, Lund University, Sweden. “It can be used for diagnosis, follow-up, and treatment evaluation of alcohol use disorders” (Figure 2).
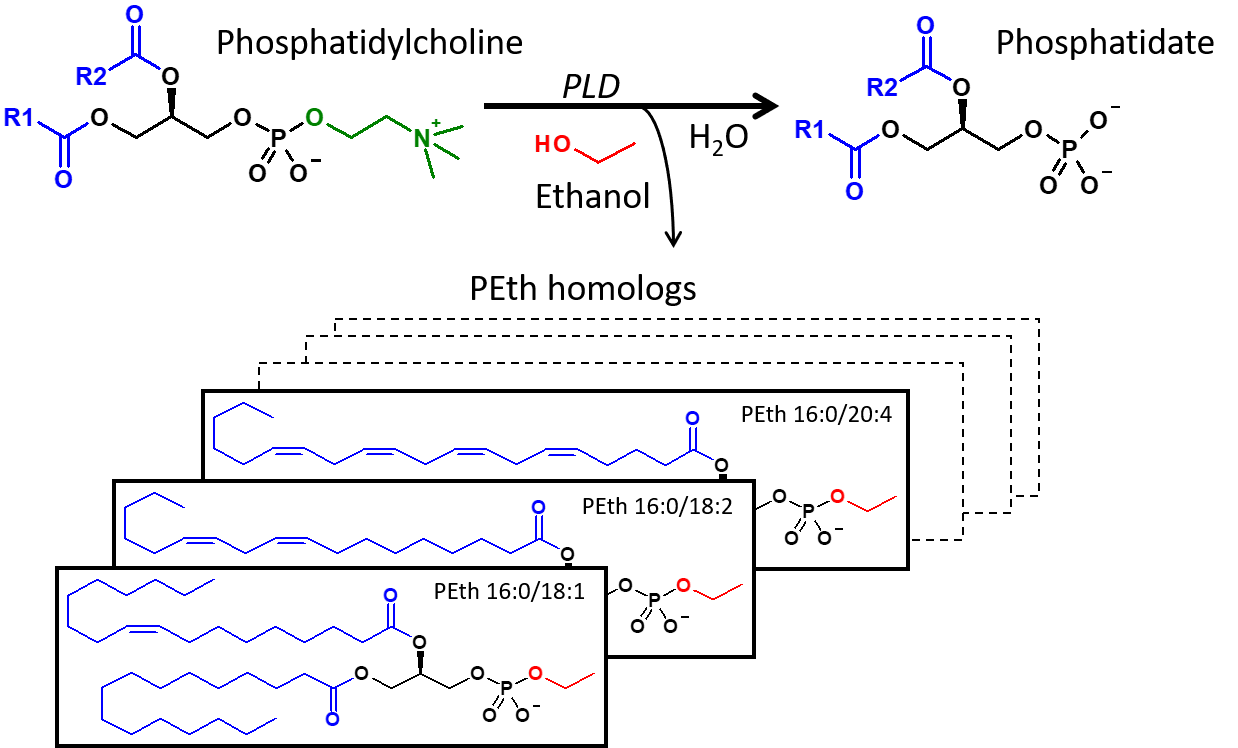
Figure 2. At normal conditions, phospholipase D (PLD) hydrolyzes phosphatidylcholine (PC) into phosphatidate and choline, but in the presence of ethanol, PLD catalyzes the transphosphatidylation of PC into the corresponding phosphatidylethanol (PEth) homolog. (Source: Isaksson et al, JALM, AACC 2018).
Isaksson noted that over the last few years, the number of samples sent to his lab to test for PEth had steadily increased, prompting him to develop a high-throughput analytical screen that was also cost-effective and reliable. “We needed an analytical method with higher throughput and analytical/clinical sensitivity than our previous HPLC-method which was based on an evaporative light scattering detection,” he explained. “Due to the large number of samples and the manual labor involved, we wanted to create a sample workup that could be robotized.”
Isaksson’s team was able to come up with a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method to determine the presence of PEth in under 3 minutes, published last year (3). During development, the challenge was to convert the sample work-up (which consisted of a protein precipitation/extraction step) to an automated procedure with sufficient analytical sensitivity and selectivity. The Lund University researchers succeeded in their efforts and have proven that their method is well-suited for a high-volume clinical laboratory that receives > 50,000 clinical samples/year.
“Our method uses an automated sample workup with only minor human intervention, and it is robust and has a high capacity. External quality samples have shown a standard deviation of < 2 SD from the method mean for all samples, since the start of the control program in 2013,” Isaksson enthused. However, he added that standardization of the method is not complete as it requires access to certified reference material, which is currently unavailable.
References
1. Imaduwage, K. P.; Lakbub, J.; Go, E. P.; Desaire, H., Rapid LC-MS Based High-Throughput Screening Method, Affording No False Positives or False Negatives, Identifies a New Inhibitor for Carbonic Anhydrase. Scientific Reports 2017, 7 (1), 10324.
2. Viel, G.; Boscolo-Berto, R.; Cecchetto, G.; Fais, P.; Nalesso, A.; Ferrara, S. D. Phosphatidylethanol in Blood as a Marker of Chronic Alcohol Use: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences 2012, 13(11), 14788–14812.
3. Isaksson, A.; Walther, L.; Hansson, T.; Andersson, A.; Stenton, J.; Blomgren, A. High-Throughput LC-MS/MS Method for Determination of the Alcohol Use Biomarker Phosphatidylethanol in Clinical Samples by Use of a Simple Automated Extraction Procedure—Preanalytical and Analytical Conditions. The Journal of Applied Laboratory Medicine 2018, 2 (6) 880-892.