Measuring Millions of Protein–Protein Interactions Simultaneously

Complete the form below to unlock access to ALL audio articles.
Testing a library of drug candidates against just one target, is both tedious and costly. In addition, for complex diseases such as cancer, many targets must be tested to determine efficacy, specificity, and toxicity. To combat this challenge, A-Alpha Bio has harnessed the power of synthetic biology and next generation sequencing to devise a more efficient way to develop drugs.
I recently had the pleasure of speaking with David Younger, PhD., Co-founder and CEO of A-Alpha Bio, to learn more about AlphaSeq – a platform designed to measure millions of protein–protein interactions simultaneously.
Laura Lansdowne (LL): Could you tell us more about A-Alpha Bio? How did the company come about and what is your vision for the company?
David Younger (DY): A-Alpha Bio aims to improve the way that drugs are developed using cutting-edge synthetic biology and next generation sequencing technology. The idea for the company began with an observation that wet-lab tools used for discovering and optimizing therapeutics are often extremely limited in throughput. In response, we developed an approach that uses genetically engineered yeast cells to measure millions of protein interactions simultaneously at a library-on-library scale. My co-founder, Randolph Lopez, and I spun A-Alpha Bio out of the University of Washington’s Institute for Protein Design and Center for Synthetic Biology in 2017. We are focused on partnering with pharmaceutical companies to discover and optimize high-impact multi-target therapeutics.
LL: What is AlphaSeq? Could you delve into the workings of this innovative platform
DY: AlphaSeq is a platform for measuring millions of protein–protein interactions simultaneously and enables the development of new high-impact therapeutics. The AlphaSeq platform involves the genetic reprogramming of yeast cells such that interactions between proteins attached to the cell surface cause cells to physically stick to one another and fuse to combine their genetic information. We then use next generation sequencing to quantitatively measure the interaction strengths of millions of protein interactions in a single step. With this technology, we can characterize interactions between a library of biologics and a large panel of target proteins to simultaneously evaluate specificity and multi-target affinity. We can also characterize small-molecule protein modulators, both antagonists and agonists, on whole protein interaction networks.
In other words, we are reprogramming yeast mating. Wild-type S. cerevisiae (baker’s yeast) have two haploid mating types, MATa and MATalpha. When mixed, a protein on the surface of MATa cells, Aga2, binds strongly to a protein expressed on the surface of MATalpha cells, Sag1. This interaction binds the two cell types together, like Velcro, even when cells are swirling around in a turbulent liquid culture. After sticking, the two cells “mate”, or fuse to form a diploid cell. We began by ‘neutering’ the yeast cells with the knockout of these native “Velcro” proteins. Next, we display proteins that we are interested in studying on the surface of a MATa cell and a MATalpha cell.
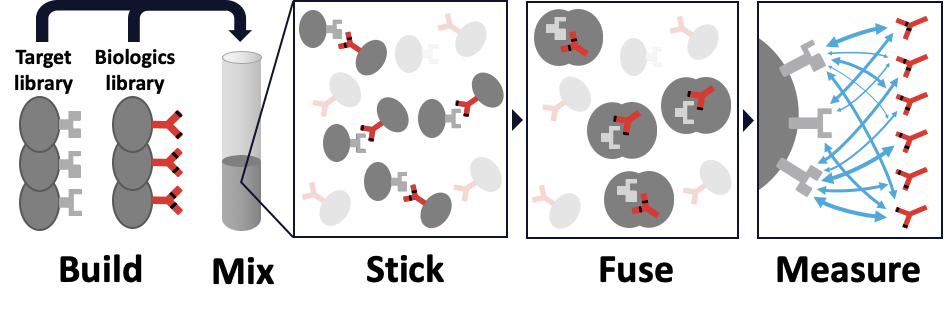 An overview of the AlphaSeq technology.
An overview of the AlphaSeq technology.
When mixed, we can evaluate whether or not the proteins interact by observing how many diploid cells are formed. This reprogramming of yeast mating to measure protein interactions is the basis for the AlphaSeq platform. We can then extend this method by combining many MATa strains, each expressing a unique protein, and many MATalpha strains, each expressing a unique protein, into a single AlphaSeq assay. Strong interactions produce many diploids and weak interactions produce few. We use next generation sequencing to count the number of diploid cells formed from each interaction and use the relative abundances to determine relative protein interaction strengths for all pairwise interactions. With this approach, we can measure protein interactions at a “library-on-library” scale.
LL: What would you consider to be the key challenges faced by drug discovery researchers, and what advantages are there to adopting the AlphaSeq approach?
DY: By enabling researchers to test millions of protein interactions at a library-on-library scale, AlphaSeq allows pharmaceutical companies to develop treatments for difficult disease targets with a significantly quicker and less expensive process than they were previously able to achieve and enables them to develop new therapeutics that would previously have been infeasible.
Today, pharma companies are only able to test a library of drug candidates against one target at a time, which is both time-intensive and costly. This is particularly true for disease classes in which many targets must be considered for efficacy and specificity – such as immuno-oncology and infectious disease. Currently, to discover an effective biologic, a drug discovery researcher would screen a large library of candidate biologics against one target at a time. If they want to hit multiple proteins or avoid likely off-target interactions, each of those screens must be performed iteratively. AlphaSeq allows them to screen against potentially thousands of on- and off-targets at once.
Discovering small molecules that modulate protein interactions is also very inefficient when only one protein interaction is measured at a time. For small molecules that disrupt or enhance particular protein interactions, AlphaSeq makes it possible to evaluate the effect of a compound on a whole protein interaction network rather than a single protein-protein interaction.
LL: Drawing upon your experience as a research entrepreneur, is there any advice you could give to those in the field that are thinking of setting up a business?
DY: I could go on and on about my experience starting A-Alpha Bio and all of the lessons that I have learned, so I’ll do my best to pick just a few. I have spoken to many researchers in the Seattle area about their interest in starting a company based on their thesis or postdoctoral work and the biggest barrier seems to be a psychological one – people don’t think they have all of the skills and knowledge that they need to start a company because they don’t have business experience. But the reality is that no one has all of the answers when they start a company. Scientists are trained to be comfortable with uncertainty and to test hypothesis with unbiased experimentation. So, my advice is to treat company building like a scientific problem. Start by identifying your hypothesis – who is your customer? What value are you adding? And then go out and test them.
My second piece of advice is to talk to as many technical founders as you can to learn from their successes and failures. There is no reason to reinvent the wheel and people in the startup community tend to fully subscribe to the “pay it forward” mentality.
And finally, although it’s not advice, I’ll add that starting A-Alpha Bio has been far more fun than I could have ever imagined. I work collaboratively with an incredible team of scientists to solve fascinating and high-impact problems. I share our science and company vision with all sorts of interesting people in pharma, venture capital, the media, and the general public. And I’m constantly learning and being challenged in new ways. Yes, the days can be long and there are stressful moments, but now I can’t imagine doing anything else.
David Younger was speaking to Laura Elizabeth Lansdowne, Science Writer for Technology Networks.


