Above the Genome: How Epigenetics Influences Cancer and Obesity

Complete the form below to unlock access to ALL audio articles.
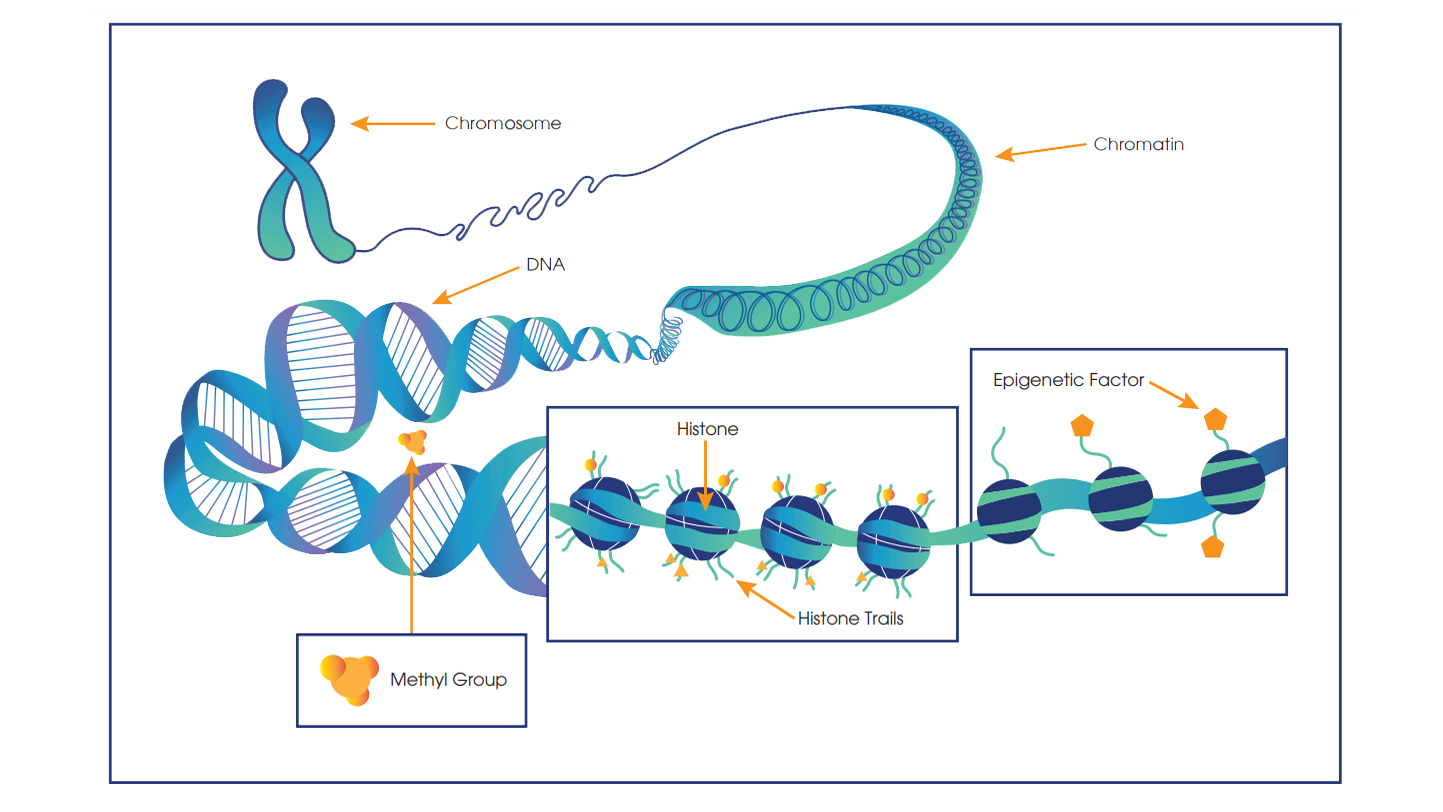
Truly understanding how our DNA influences our lives and health sometimes means moving past the DNA sequence. Epigenetics encompasses a group of heritable modifications without alterations to the DNA sequence itself, which is the domain of genetics. Epigenetics derives its root from the Greek prefix “epi” meaning “above” or “over,” and when linked with “genetics,” “epigenetics” literally translates to “on top of genetics,” alluding to an added layer of heritable characteristics in addition to genetic inheritance. Epigenetic changes can occur by a variety of mechanisms, such as modification to DNA bases, e.g., by methylation of cytosines, alterations to histone proteins, e.g., by methylation or acetylation, and a general remodeling of chromatin ultimately leading to changes in gene expression profile.
Deregulation of the epigenome can have far-reaching impacts on our health and consequently epigenetics has been increasingly appreciated in recent decades in a staggering number of diseases. Although cancer tops the list, epigenetics has featured in obesity, metabolic syndromes, and diabetes, aging, cardiovascular disease, kidney and liver diseases, autoimmunity, such as rheumatoid arthritis, allergic diseases, and inflammatory diseases such as Crohn’s disease and inflammatory bowel disease. Epigenetics also plays a role in depression, neurological and neurodegenerative illnesses including Alzheimer’s disease.
Cancer capitalizes on epigenetic deregulation
Aberrant epigenetics is a central feature of numerous cancers and has long been an intensive area of study. Epigenetics can alter the proliferation, cell-cycle progression, metabolism, and metastatic potential of cancer cells, ultimately driving tumorigenesis and disease progression. Several drugs targeted at epigenetic deregulation to treat cancers are approved by the FDA, such as histone deacetylase inhibitors (iHDACs), alkylating, and demethylating agents, in addition to drugs in preclinical and clinical development.
Although much progress has been made, particularly for blood cancers, epigenetic deregulation continues to emerge as a problem in several forms of cancer. Recently, several types of brain tumors were found to be epigenetically deregulated, such as mutant isocitrate dehydrogenase 1/2 (mIDH1/2) low-grade glioma and pediatric high-grade diffuse intrinsic pontine glioma (DIPG). mIDH1/2 glioma is characterized by hypermethylation to both DNA and histones due to altered cellular metabolism. This restructuring of the methylome, which is the pattern of histones and DNA bases modified by methylation across the entire genome, is ultimately believed to drive tumorigenesis and drugs targeting mIDH1/2 enzymes are in clinical development.
DIPG is a fatal childhood cancer with an especially poor median survival of only 11 months. Conventional therapies produce only modest improvements in patient outcomes and new treatment avenues are sorely needed. Molecular profiling of DIPG tumors recently uncovered the first ever mutation to a histone protein, histone 3 (H3), most frequently at amino acid K27 to M (H3K27M). “This was a surprise,” recalls Dr. Cynthia Hawkins, Neuropathologist and Senior Scientist, The Hospital for Sick Children, Toronto. “Although alterations involving the PRC2 complex, in particular its EZH2 domain, had been previously described in other cancers, this was the first reported mutation in a histone gene.” PRC2, or polycomb repressive complex 2, is a histone methylating enzyme, i.e., methyltransferase, also involved in regulation of the epigenome. Overexpression or mutation to PRC2 had been known to drive several types of cancer.
Understanding the mechanism driving epigenetic deregulation can shed light on potential therapeutic targets and is an intensive area of research in mutant H327M DIPG. The H3K27M mutation alters the K27 residue, an amino acid that is frequently modified by methylation. “Mutant H3K27M histone is expressed as a small percentage (~5-15%) of total H3 because there are multiple genes encoding H3, not because the wild-type allele is expressed more strongly,” explains Dr Hawkins. However, this small fraction of expressed mutant H3K27M drives a net global hypomethylation. “The mechanism of this effect is believed to occur by inhibition of EZH2, the methyltransferase domain of PRC2. PRC2 is thought to get locked on the mutant methionine residue, preventing it from moving along the chromatin to methylate adjacent wild-type histones, which reorganizes the methylation status at H3K27 across the genome with some areas showing loss and others gain of trimethylation.” Overall, an altered methylome that has several downstream consequences for DIPG tumors.
What does this spell for a potential treatment? “Targeting the mutant histone directly has proven difficult,” responds Dr. Hawkins. “There is an open trial using an immunotherapy-based strategy to target cancer cells; however, there have not yet been dramatic responses to this strategy. Other trials targeting more general epigenetic mechanisms are being considered based on preclinical data and are underway such as HDAC inhibitors, EZH2 inhibitors, and BMI-1 inhibitors.”
Omics cast light on obesity
Obesity is a multifactorial disease, a product of environmental factors such as dietary intake and intrinsic ones such as genetic predisposition. Although early lines of investigation focused on dietary consumption as a primary reason, the relatively stable ratio of obese to lean people in the same obesogenic environment suggested variation and predisposition among individuals. Identification of these factors has been made easier by the rise of Omics system-wide approaches, which have revolutionized the study of obesity. For instance, genome-wide association studies (GWAS) have identified key loci pertinent to genetic predisposition to obesity.
However, the loci uncovered by GWAS studies could not account for all the heritability observed in obesity, which has led investigations into potential epigenetic contributions to obesity. Earlier studies examined specific, individual loci, but epigenome-wide association studies (EWAS) have identified a greater number of loci. “The effects of epigenetic modifications, such as DNA methylation, on the development of obesity has been demonstrated in experimental research, such as animal studies, as well as in humans,” explains Professor Lu Qi, Professor at Tulane University School of Public Health and Tropical Medicine and Adjunct Professor at Harvard T.H. Chan School of Public Health. “Recently, genome-wide studies have provided convincing data to link DNA methylation to human adiposity (i.e., BMI, body mass index), likely through a causal pathway.”
Many investigations into epigenetic changes and obesity have centered on DNA methylation, but some studies have examined the impact of other transcription regulators, such as microRNAs (miRNAs) on obesity. Moreover, the majority of work has examined the impact of epigenetics early in human development. “It is believed that dietary or lifestyle changes more likely affect epigenomic changes (e.g., DNA methylation) during a developmental window, such as during the prenatal period or early childhood,” Professor Qi elaborated. “It is feasible that dietary or lifestyle changes may also affect obesity-related epigenomic changes in later life, but the supporting evidence is currently lacking.”
So how might adults manage their weight or how can obesity be treated? The multitiered contributions to obesity, from personal food intake to genetic, epigenetic, and metabolic makeup advocate a “personalized” approach to nutrition to help individuals manage body weight. “Personalized dieting is essentially a similar concept to precision medicine; both emphasize that intervention or treatment should take into account individual variability, which is determined by intrinsic factors such as genomic and epigenomic variations, among others,” Professor Qi explains. “I think in the future, we will be able to identify various subgroups of people who may respond differently to dietary interventions, according to their genomic, epigenomic, and other profiles. On this basis, precision interventions may manage obesity more effectively.”
Roadmap for the future
The rise in our understanding of the epigenetics of disease has been facilitated by a suite of sophisticated methods to study the epigenome. Many have become almost routine, such as whole-genome bisulfite sequencing (WGBS) and methylation microarrays to study methylated DNA loci, chromatin immunoprecipitation with sequencing (ChIP-seq) for histone modifications, and assay for transposase-accessible chromatin using sequencing (ATAC-seq) for chromatin accessibility, to name just a few. In addition, novel methods continue to emerge, including single-cell techniques to study population heterogeneity. Such improved methods of epigenomics analysis will help uncover mechanisms of epigenetic deregulation in disease, identify potential epigenetic therapeutic targets, and assess novel drugs targeted to epigenetic deregulation, thus paving the way for the treatment of epigenetic diseases.



