Molecular Science and Art With Professor David Goodsell
Complete the form below to unlock access to ALL audio articles.
Perhaps perceived as distinct lines of enquiry, expression and study in the past, the fields of art and science are now more widely accepted as being deeply connected. While their tools and mediums differ, the overarching objective is to communicate an understanding of the world around us.
Albert Einstein allegedly said that “the greatest scientists are artists as well”. Interpretation of the word “greatness” is, of course, subjective. But if we consider the most prestigious accolade in the scientific world – the Nobel Prize – as one measure, Einstein’s point has merit. In Arts Foster Scientific Success, Professor Robert Root-Bernstein, a scientist, humanist and artist at Michigan State University and his wife Michele Root-Bernstein, a scholar, writer and poet analyzed the artistic vocations of Nobel Laureates from the period of 1901–2005. Their data suggested that Laureates were 2.85 times more likely than the “average scientist” to possess an artistic or crafty vocation. Now, Nobel Prizes are few and far between in the career of most scientists, but the paper presents an interesting exploration of the qualities that creative endeavors might afford a researcher.
It appears the relationship between art and science might be likened to something symbiotic in nature. The artistic process may provide an outlet for self-expression that yields inspiration, encouraging the consideration of alternative perspectives. Similarly, artwork can provide a visual depiction of scientific concepts, allowing the comprehension of complex, perhaps even microscopic phenomena.
In the minds of many, Professor David Goodsell embodies the intertwining of science and art in the 21st century. A structural biologist-turned molecular artist, Goodsell holds joint appointments across two prestigious laboratories. At the Scripps Research Institute, he is a professor of computational biology in the Department of Integrative Structural and Computational Biology. Over at Rutgers University, he creates materials for outreach and education at the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB), a global resource archiving 3D structure data for large biological molecules, and the home of Molecule of the Month.
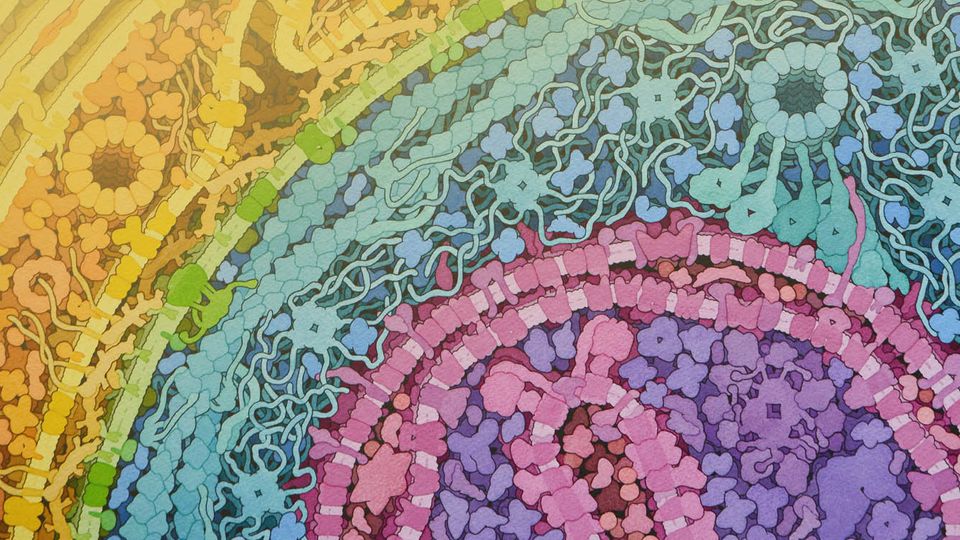
 Credit: David Goodsell.
Credit: David Goodsell.
Structural biology involves studying the structure and dynamics of biological molecules, exploring how this relates to function, and how function can be perturbed by structural changes. In Goodsell’s own words, “the power of SciArt has perhaps its strongest manifestation in structural biology, where the things we study are particularly amenable to visual representation.” Throughout his almost three decade-long career, Goodsell has created hundreds of pieces of artwork depicting the beautiful intricacies of the delicate molecular world. His portfolio includes paintings of molecules that are central to life – such as acetylcholinesterase, collagen and hemoglobin – and heinous pathogenic invaders, like Ebola or HIV. He describes his cellular landscapes, created using vibrant shades in watercolor, as “simulating a view of the cellular mesoscale, taking a portion of the cell and depicting its molecular composition”.
A major contributor to scientific literature, Goodsell has also authored four books including: 1. The Machinery of Life 2. Our Molecular Nature: The Body's Motors, Machines and Messages, 3. Bionanotechnology: Lessons from Nature and 4. Atomic Evidence: Seeing the Molecular Basis of Life. His passion and commitment to science, art and education are widely appreciated; most recently he was presented with the 2022 Fankuchen Memorial Award by the American Crystallographic Association as recognition for “contributions to crystallographic research by one who is known to be an effective teacher”. The press release accompanying the award acknowledges Goodsell’s “extraordinary skills as a communicator of science”.
Goodsell describes himself as a scientist, first and foremost – the goal of his art is to “create imagery as a tool for science”, in his opinion. For issue 20 of The Scientific Observer, Technology Networks had the privilege of interviewing Goodsell – who is as humble as he is talented. We spoke about his early experiences in science and art, the creative process behind his stunning imagery and pursuing a career in this space.
Molly Campbell (MC): You started painting in your childhood but chose to pursue science as a career. Can you talk to us about this decision?
David Goodsell (DG): My first interest was in natural history – plant collections and insect collections – and from there the interest evolved to science more broadly. My father was an aerospace engineer, so I largely inherited a passion for science and technology from him, but my whole family could be described as “arty”. My grandfather was a watercolorist, and he started teaching me how to paint with watercolor from an early age.
I always a “science nerd”, and I suppose I knew that – from a professional standpoint – I was going to be a scientist; I’ve just always created art on the side for fun and to stay sane. When I entered graduate school, that’s when I started to really bring art into my daily scientific work. Ever since then, I’ve used art to “serve the science”. I was lucky that, while in grad school, computer graphics hardware started to become affordable. The department that I joined at the University of California Los Angeles (UCLA) had purchased really cool AI and computer graphics software that nobody knew how to use. I got to experiment with writing programs and developing methods that supported the science that I was doing. Really, it allowed me to have some artistic freedom to play around. UCLA was a nice example of where the artistic approach helped me to understand the science and was appreciated by other scientists that saw the value in doing that.
MC: Did you enjoy your PhD experience?
DG: It was wonderful. I had always dreamed about doing real research, and suddenly, I found myself in a lab where, for the first time, I could design an experimental approach based on a problem and come up with the tools required to solve that problem.
My advisor Professor Richard Dickerson was an amazing follow – a classic example of a gentleman scientist. I’m amazed at how much everything I’ve done thereafter is styled on his approach. He was able to teach me the basics of crystallography and how to interact with colleagues in a collegial way, but he also had a long history of writing influential chemistry books and had worked with the artist Irving Geis for many years. They created several general interest books on protein structure, and Dickerson was very amenable to me working on these visual methods alongside my core research. The environment was perfect for what I wanted to do.
 Geis illustrates the hemoglobin molecule as four symmetrically arranged myoglobins. Since it is responsible to the transport of oxygen, it can change from an oxygen-binding configuration to an oxygen-releasing configuration in response to the demand for oxygen. Credit: Illustration, Irving Geis. Used with permission from the Howard Hughes Medical Institute (www.hhmi.org). All rights reserved.
Geis illustrates the hemoglobin molecule as four symmetrically arranged myoglobins. Since it is responsible to the transport of oxygen, it can change from an oxygen-binding configuration to an oxygen-releasing configuration in response to the demand for oxygen. Credit: Illustration, Irving Geis. Used with permission from the Howard Hughes Medical Institute (www.hhmi.org). All rights reserved.

David Goodsell and Richard Dickerson exploring a structure of DNA bound to the trial antitumor compound netropsin, using the interactive display of an Evans and Sutherland (E&S) MultiPicture System and custom software. Credit: The University of California, Los Angeles.
MC: Do you remember the first scientific project that you looked at through an artistic lens?
DG: For sure! At the time I was working with Dickerson, using X-ray crystallography to study little pieces of DNA to understand how anti-cancer drugs bind to them. We would get the crystallographic maps and print them out on acetate transparencies, stack them up with sheets of plexiglas and measure everything out by hand. Then, a new graphics system – called the multi-picture system – was introduced that allowed you to do a lot of this work interactively on the screen, but there wasn’t a good software for it. Some of the first projects that I completed involved taking the structures and the crystallographic maps that we were determining in experimental work and coming up with graphical ways to look at these new computer graphic systems.
MC: What is it about solving structures that interests you?
DG: I distinctly remember the first electron density map that I obtained from one of the DNA structures. When we put it on the screen, I thought to myself, “here I am seeing these individual atoms”. That was a big point in my training – the realization of “we can actually use these methods to see these biomolecules”. Everything was built from that. I think I’ve always been interested in structural biology and that probably comes from my early interest in natural history. I like to see things and classify them and understand variability. It’s a very visual and tangible approach to see something’s structure and figure out how it works.
MC: You now hold two appointments across the Scripps Research Institute and Rutgers University. From the early graduate experiences that you talk about, how has your research focus evolved to what it is now?
DG: I’ve always tried to split what I do in half – research being one half and the outreach work being the other. When I started at Scripps, all my work focused on the research; I did all of the artwork, illustration, education and outreach in my spare time.
As my career has progressed, and I've become known more for the outreach and artwork, I've found ways to support myself with that. That's where the RCSB comes into the picture. The Molecule of the Month project started as a development of some software used for exploring the structures of molecules, and I was writing little stories to accompany that software. It spiraled from there and turned into a way for the RCSB to popularize the structures that are available in the archive, and to expand the user community to populations that might not understand how structure data can be used in their own research or lives.
So, the trajectory of my career very much started from being a scientist. And then as my reputation increased surrounding my artwork, I was able to increase my work in that area and that’s when the Rutgers appointment happened.
The Molecule of the Month
Launched in January 2000, the RCSB PDB “Molecule of the Month” is part of the PDB’s educational portal. Authored by Goodsell, it presents short overviews of molecules from the PDB, outlining an introduction to the structure of the molecule, its function and its relevance for the life sciences and beyond.
As an open-access resource, Molecule of the Month has proven to be a critical educational tool, as revealed by a 2019 survey by the RCSB PDB, which found that, of 339 respondents, ~50% use the column for teaching, ~60% work within a college or university and ~78% work in biology and biochemistry. An editorial published by Goodsell and colleagues on the project’s 20th birthday confronted the question: after 20 years, aren’t you running out of molecules? “New biostructural methodologies, such as cryoelectron microscopy and time-resolved crystallography with X-ray free-electron lasers, are opening up entirely new fields of structural study and providing structures of ever-increasing complexity for inclusion in the column,” the authors responded.
MC: Your artwork portfolio includes watercolor creations depicting the inside of a cell environment, drawn at-scale. This sounds incredibly complex – can you talk to us about how you create these images, and how you ensure that they are precise?
DG: There really are two types of ways that it can be approached. One is immersive in style, like you’re taking a voyage in to the cell, and you find yourself in the middle looking around at the environment that surrounds you.
The alternative approach – which I use – is where you imagine you have taken a cell and “clipped it”, so you’re looking at a section in front of you at a bit of a distance. It’s like a flat cut into the cell. I use this approach because, firstly, it allows me to show a panoramic portion of the cell, and secondly, because I can draw everything at the same scale, and you can compare the individual parts from a size perspective.
It’s not as dramatic as the immersive approaches, but to me there can be too many distortions with those – I lose track of what I’m trying to show. Of course, this will likely change soon as animation is becoming much more accessible, easier to use and increasingly immersive – take virtual reality for example. People in my lab are experimenting with the new animation approaches to attempt to create the “immersive” experience of a cell, but I do think it is very much “up in the air” as to how you can understand scale relationships in such environments.
About being precise: I always try to capture the current state of knowledge in these illustrations. Most often, half of my time is spent researching a topic before I even start sketching. When I started, this process was laborious, involving lots of time in the library. Today it’s much easier, since we have wonderful online resources like the RCSB PDB, UniProt and PubMed that puts all this information at our fingertips.
MC: Many people describe themselves as a visual learner. What is your perspective on learning visually and how people might use your images to understand concepts?
DG: I’ve always described myself as a visual person and a visual learner – if I can see it, I can understand it better.
One important aspect of the cell pictures is that they offer a view of a cell that isn’t available experimentally, or at least wasn’t until very recently. Crystallography methods give you a very detailed structural understanding of a molecule, while microscopy looks at whole cells but doesn’t always allow you to see the individual molecule. The images that I create synthesize the information to create a view that really isn’t available from experimental methods.
That’s changing now with cryo-electron tomography, which is routinely showing things like individual ribosomes inside cells. But to get the full view, showing every molecule, simulated images are still the only approach.
I think offering a scientist a visual concept of just how crowded cells are gives them a touchstone when they’re thinking about their experiments. It’s a view of cells that really wasn’t available before I started creating the pictures. People knew they were crowded, but they didn’t really internalize that when they were thinking about their work. I’d describe that as the biggest impact of my pictures.
Also, with the RCSB PDB, I’ve worked very hard to create a style that makes the individual protein structures accessible and feel familiar. I use a cartoon-like approach that is colorful and hopefully more interpretable than the complex diagrams you might see in the more technical literature.
MC: What are some of your favorite projects that you have worked on throughout your career?
DG: The paintings that I do of cellular interiors are something that I still enjoy doing immensely. When I started working on those in my postdoc, I treated it as a challenge to myself. It felt like a treasure hunt. At the time, I didn’t know whether there would be enough information to really create an accurate picture that shows all the molecules inside of the cell. I had to spend so much time in the library gathering the information I needed and trying to come up with the visual methods to make that complex space be interpretable. When I completed my first picture, I think that was probably the most rewarding period of my creative career so far.
Another period of my career that was really exciting was during my graduate work in the early days of computer graphics. The whole field was super exciting because nobody knew how to do anything, so everyone was making it up as they go, so it was a time of trying new representations, new approaches to lighting and experimenting with visual methods to make these details more accessible and interpretable.
MC: I suppose that, with the nature of scientific discovery, your work is never truly finished. New data is published constantly, which changes our understanding of certain concepts. How do you work with that?
DG: You must work with what you have available – for me, that’s the information that is published, and my ability to find that information. I’m lucky that most researchers don’t pick out tiny details and say “actually, that’s incorrect”, rather they see it as a larger, general synthesis of the information available.
I have been creating these pictures for almost 30 years now. For me, looking back at the pictures I’ve made in the past – take the half a dozen Escherichia coli structures I’ve drawn, for example – I’m seeing those pictures as snapshots of the information that I could find at the time. It shows that science is a living process that continues to change and progress. I don’t worry too much about the fact that the pictures will go out of date, largely because I often have an application for them at the time they are made, so the picture needs to capture what people are thinking or know today.
MC: What happens if, at the time of creating a picture, you don’t have all the information that you need?
DG: What I do is very much dependent on how the image will be used. I don’t like to create images where I have to make a lot of the content up. When people ask me to do that, I tell them that science isn’t up to it yet.
MC: Your work is used far and wide as an educational tool, to accompany journal research articles and beyond. How does it make you feel to know you are having a major impact on the world’s understanding of science?
DG: It’s exactly why I do this, so that I can help people see the cells in the way that I am seeing them. Foremost, I make the art for myself because I am so interested in seeing these views, but it is wonderful to see other people visualizing cellular processes or molecules the way that I see them, and also building on them – adding whatever information they can to the picture.
For instance, the cell images are commonly used in textbooks, most often presented at the beginning of the book to say “look, cells are really crowded places”. Then the rest of the textbook focuses in on these individual elements to breakdown how they are working to create this holistic picture. It’s incredibly rewarding.
I am so lucky to be able to work with the RCSB PDB and make all the images freely available with Creative Commons. The only reason that I can do that is because the RCSB PDB offer me the support I need, which means they truly see the value of the artistic approach in science.
MC: Can you tell us about “CellPAINT”, what it is and how you hope it will be used?
DG: As part of our work on creating 3D models of whole cells, I wanted to create an easy-to-use method for people to use to create their own cellular environment pictures. Two talented people in the lab, Dr. Ludovic Autin and Adam Gardner, ran with the idea and created CellPAINT. It works like a familiar digital painting tool, but instead of brushes, you pick molecules from a palette to paint into the scene. We’re currently putting the finishing touches on a virtual reality version of it that will make it easier to build similar 3-dimensional scenes.
MC: Pursuing a career in SciArt might not seem an obvious path for someone completing an education in science, or art. What advice can you offer to somebody that reads our interview and thinks they would like to work in this space?
DG: I get a lot of enquiries asking how to follow a similar career path to mine. I always tell people that when I look at my community of friends and colleagues, there are really two pathways they have taken to get where they are.
The first is the pathway that I took. I pursued a training in science, which makes science my primary profession, and then I do artwork in support of that. To me, this is the most realistic path to take to be able to make a living in this space. The jobs available in science are more defined, right? And the scientific community is hungry for this type of imagery, but it’s not always willing to pay for it.
The other path that I see people follow involves diving headfirst into the art world first. There are some beautiful schools around the world to study scientific art, and in the United States, there are even medical illustration colleges such as the Association of Medical Illustrators and the Guild of Natural Science Illustrators.
These institutions can support people who wish to forge a career in scientific and medical illustration. For individuals that choose that route, I see doors open for a variety of different types of jobs – as textbook illustrators, freelance artists working with drug companies, in museums, etc. Finding that kind of work might not be as obvious a route as following a career into research but can be equally rewarding.
MC: Who else in the molecular artwork space inspires you?
DG: I am a voracious consumer of scientific art, so I draw inspiration from many, many sources. Irving Geis, of course, is the father of molecular illustration and launched me on my path. Currently, there are several people that I think of as competitor/collaborators: friends who are working in much the same space as I do. These include Professor Drew Berry, Dr. Janet Iwasa, Dr. Gael McGill and Dr. Beata Mierzwa.
MC: What would you describe as being the greatest challenge in your work?
DG: I need to integrate many types of data (atomic structures, proteomics, microscopy, etc.), and it's always a challenge to deal with the current heterogeneous data environment. Typically, each of these types of data has its own database, format, search tools, etc. In some cases, online databases are not available, so I need to go directly to the scientists. This challenge is getting better and better – 20 years ago, none of these web-based information sources were even available. But it still feels like a treasure hunt every time, as I try to gather the information that I need to support the illustrations.
Professor David Goodsell was speaking to Molly Campbell, Senior Science Writer for Technology Networks.



