Understanding the Lytic Cycle – What Are the Steps?

Complete the form below to unlock access to ALL audio articles.
Bacteriophage (phage) are viruses that specifically infect bacteria. They have a two-phase lifecycle, residing in a dormant state within the host genome (lysogenic cycle) or hijacking the host cellular machinery for their own replication (lytic cycle). Here we will explore the important steps of the lytic cycle.
Contents
- Phage attachment
- Bacterial cell entry
- Phage replication
- The birth of new phage
What is the lytic cycle?
Whilst the ultimate outcome of the lytic cycle is production of new phage progeny and death of the host bacterial cell, this is a multistep process involving precise coordination of gene transcription and physical processes. The phage must identify a susceptible and suitable host bacterial cell to which it is able to attach. Following introduction of phage genetic material into the cell, the host genome is destroyed, and the phage utilizes the host cellular machinery to make copies of its own genome and synthesize the structural components. New phage are assembled before the host cell finally lyses and the phage progeny released into the surrounding environment to find new target cells.
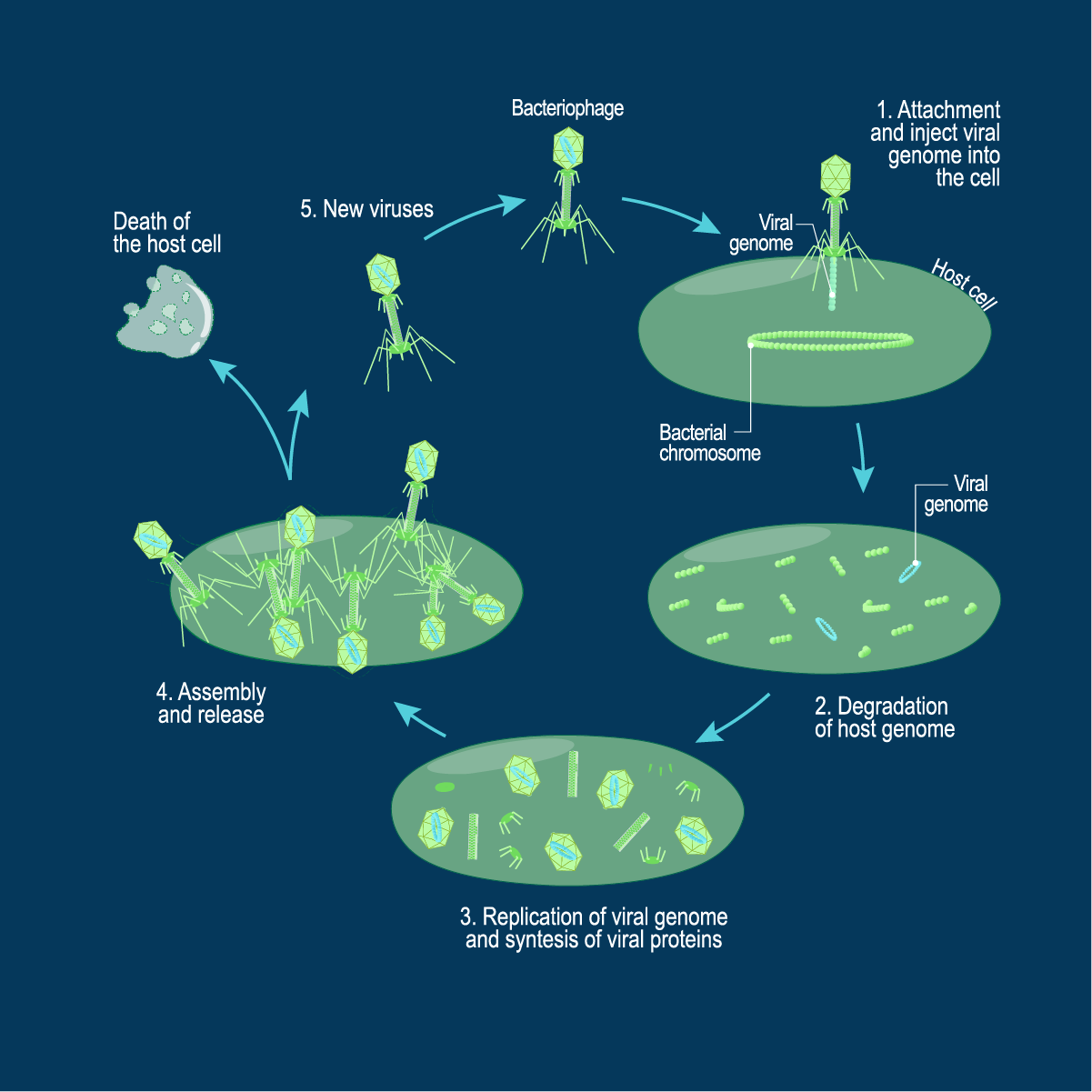 Credit: Technology Networks
Credit: Technology Networks
Lytic cycle steps
- Phage attachment
In order to enter a host bacterial cell, the phage must first attach itself to the bacterium (also called adsorption). Initial contact between phage and bacterium often happens through random collisions and initial binding is reversible. Not all bacteria-phage combinations have compatible receptors and receptor binding proteins, so this is a selective process. Different phage species target a range of different molecules on bacterial cells, including polysaccharide moieties, proteins and other cell surface structures such as pili. As far as we know, these target molecules all fall under the category of bacterial cell wall components or protruding structures.
Phage receptor binding proteins are most frequently associated with phage tails but have been identified in other locations on phage. Following reversible binding, the phage binds irreversibly to the cell, not always through the same receptor as the initial binding step. This two-step binding process may be beneficial especially in cases where the irreversible binding domain is less easy to access, improving the probability of pairing with the correct target cell.
Bacteria have a number of ways to defend themselves from phage attachment which center around masking the receptors with mucoid capsule or slime layers, by producing competitive inhibitors or otherwise blocking access. Phage receptors are often identified by mutations that make the bacteria resistant to phage lysis. The Phage Receptor Database (PhReD) provides a useful repository of known receptors on host cell surfaces.
- Bacterial cell entry
Successful adsorption triggers the next stages of infection which requires injection of the phage’s genetic material into the host cell. For this the phage must penetrate the bacterial cell. Evidence has suggested that attachment and penetration are coordinated by the baseplate in tailed phage. Phage tails vary widely through nature, but the most sophisticated have a tube for delivering genetic material surrounded by a contractile sheath. The sheath contracts like a coiled spring and then on release drives the tube into the bacterial cell. In T4 phage, the entire baseplate-tail-tube complex consists of around one million atoms, making up 145 chains of 15 different proteins. The empty phage cell which is left outside the bacterium is called the ghost or doughnut.
- Phage replication
Once inside, the phage synthesizes early proteins including endonucleases and exonucleases that degrade the host genome. They are then able to use the host cell’s machinery to synthesize proteins and produce progeny. The nucleotides released may be recycled by the phage for the replication of their own progeny (e.g., T7 phage) or excreted from the host cell (e.g., T5 phage). Small chemical modifications (in the case of T4 phage, chemical modification of the viral cytidines) to the phage genome allow its genetic material to be differentiated from the host genome and prevents self-degradation during this process. Other early proteins include those required for replication of the phage genome. RNA is not broken down, so the phage may also produce inhibitors that prevent host RNA polymerase from interfering with viral polymerases during later infection. The newly synthesized phage genome produces late proteins including the capsid subunits and tail. This process can occur within minutes of the bacteria becoming infected.
- The birth of new phage
Once all the component parts have been synthesized they must be assembled into mature phage. Capsid proteins assemble to form empty heads into which condensed phage DNA is packed. The tail parts assemble independently of the head structure and the last step in synthesis is joining the filled heads to the tails to form progeny phage.
The enzymes produced by the phage gradually weaken the bacterial cell wall and eventually the bacterial cells lyse, releasing on average 100-200 phage progeny into the surrounding environment.
Due to the nature of infections, lytic phage do not alter the phenotype or genotype of the cells they infect. However, they exert selective pressure on the bacterial population, eliminating susceptible hosts and promoting the propagation of resistance, by gene transfer for example, within the bacterial population.



