English version 11 December, 2020 on EurekAlert!
Efforts to enhance the ability of proteins to resist breaking down, or ‘denaturing’, at high temperatures is one of the hottest topics in biotech. Researchers have now identified some of the principles behind how this works, potentially opening up a raft of industrial applications for designer proteins.
---------------------------------------------------------------------
Bioengineers have found why proteins that are designed from scratch tend to be more tolerant to high temperatures than proteins found in nature.
Natural proteins with high ‘thermostability’ are prized for their wide range of applications, from baking and paper-making to chemical production. Efforts to enhance protein thermostability—and to discover the principles behind this—is one of the hottest topics in biotech.
The latest discoveries, described in the Proceedings of the National Academy of Sciences on November 23, 2020, open up the possibility of lab-made proteins with even better industrial applicability.
Researchers in the relatively young field of protein design have attempted to come up with new types of proteins for myriad medical, pharmaceutical and industrial applications. Until recently, protein engineers have focused on manipulating existing natural proteins. However, these natural proteins are difficult to alter without also distorting the general functioning of the protein—much like adding a fifth wheel to a car.
To avoid this, some protein engineers have begun to build novel proteins entirely from scratch, or what is called de novo protein design.
However, this quest has its own set of issues. For example, building proteins from scratch is much harder computationally, and requires a complete understanding of the principles of protein folding—the multiple levels of how a protein literally folds itself into a particular structure.
In biology, structure determines function, much like how a key fits into a keyhole or a cog into a sprocket. The shape of a biological entity is what allows it to do its job within an organism. And upon their production by cells, proteins just fall into their shape, simply as a result of physical laws.
But the principles that govern the interaction of these physical laws during the folding process are frustratingly complex—hence the computational difficulty. They are also still largely unknown. This is why a great deal of effort in protein engineering in recent years has focused on attempting to discover these protein design principles that emerge from physical laws.
And one of the mysteries facing protein designers has been the high thermostability of these ‘lab-made’ proteins.
“For some reason, de novo proteins have repeatedly shown increased tolerance in the face of quite high temperatures compared to natural proteins,” said Nobuyasu Koga, associate professor at Institute for Molecular Science, and an author of the study. “Where others would ‘denature’, the lab-made proteins are still working just fine well above 100 ºC.”
The design principles that have been discovered so far emphasize the importance of the backbone structure of proteins––the chain of nitrogen, carbon, oxygen and hydrogen atoms.
On the other hand, these principles have also held that the tight packing of the fatty, hydrophobic (water-resistant) core of naturally occurring proteins—or rather the molecular interactions that allow them to sit together as snugly as pieces of a jigsaw puzzle—is the dominant force that drives protein folding. Just as how oil and water don’t mix, the fattier part of the protein when surrounded by water will naturally pull itself together without any need for an external ‘push’.
“Indeed, according to our design principles, protein cores were engineered specifically to be as tightly packed and as fatty as possible,” Nobuyasu Koga said. “So the question was: Which is more important for high thermostability, backbone structure or the fat and tight core packing?”
So the researchers took the de novo proteins they had designed that had shown the highest thermal stability, and began to tweak them with ten amino acids involved with the hydrophobic core packing. As they did this, they saw still folding ability and little reduction in overall thermal stability, suggesting that it is instead the backbone structure, not the hydrophobic core packing, that contributes the most to high thermostability. “It is surprising that the protein can fold with high thermal stability, even the loose core packing,” said Naohiro Kobayashi, coauthor and a senior research fellow at RIKEN.
“Hydrophobic tight core packing may not even be very important for designed proteins,” added Rie Koga, coauthor and a researcher at Exploratory Research Center on Life and Living Systems (ExCELLS). “We can create an exceptionally stable protein even if the core packing is not so optimized.”
The next step for the researchers is to further develop rational principles for protein design, especially with respect to what extent that substructures of the backbone, especially loops within it, can be altered without endangering its folding ability and high thermostability.
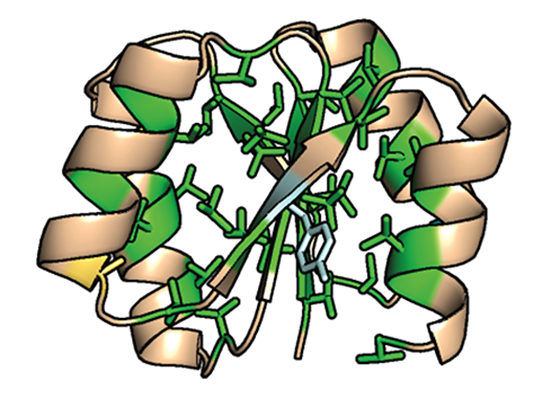
De novo protein
Structure of the de novo protein with most of the core filled with valine residues (green). Ten hydrophobic residues were mutated to smaller valine residues. This de novo protein still shows high thermal stability above 100 ºC.
Information of the paper
Authors: Rie Koga, Mami Yamamoto, Takahiro Kosugi, Naohiro Kobayashi, Toshihiko Sugiki, Toshimichi Fujiwara, and Nobuyasu Koga
Journal Name: Proceedings of the National Academy of Sciences of the United States of America
Journal Title: "Robust folding of a de novo designed ideal protein even with most of the core mutated to valine"
DOI: 10.1073/pnas.2002120117
Financial Supports
Japan Agency for Medical Research and Development (JP19am0101072)
Grant-in-Aid for Scientific Research (15H05592, 18K06152, 18H05420)
Japan Science and Technology Agency (JPMJPR13AD)
Contact Person
Nobuyasu Koga
TEL: +81-564-55-7379
E-mail: nkoga_at_ims.ac.jp
*Please replace the "_at_" with @
1977

