Genomic Analysis of L5178Y tk+/- Cells and their Induced tk-/- Mutant Colonies
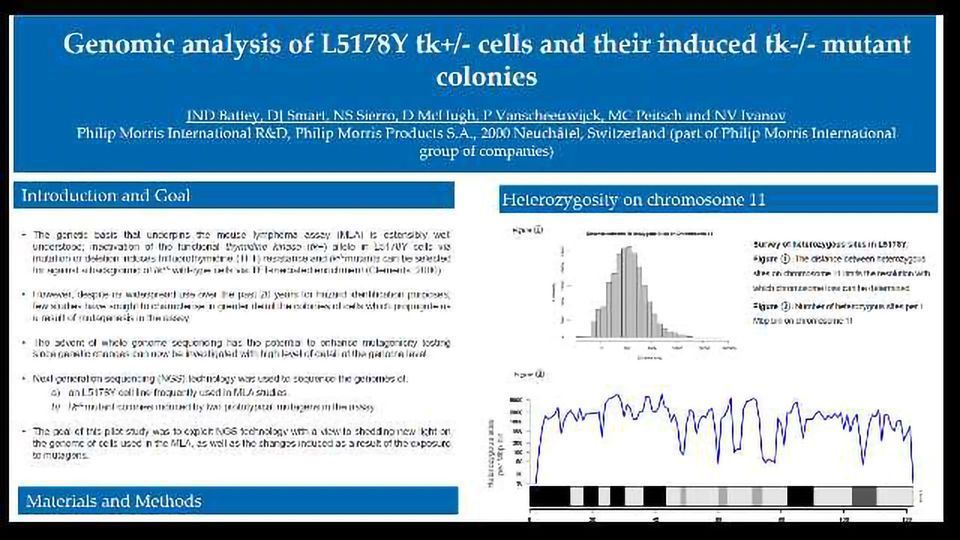
The genetic basis that underpins the mouse lymphoma assay (MLA) is ostensibly well understood; inactivation of the functional thymidine kinase (tk+) allele in L5178Y cells via mutation or deletion induces trifluorothymidine (TFT) resistance and tk-/-mutants can be selected for against a background of tk+/- wild-type cells via TFT-mediated enrichment (Clements. 2000).
• However, despite its widespread use over the past 20 years for hazard identification purposes, few studies have sought to characterise in greater detail the colonies of cells which propagate as a result of mutagenesis in the assay.
• The advent of whole genome sequencing has the potential to enhance mutagenicity testing since genetic changes can now be investigated with high level of detail at the genome level.
• Next generation sequencing (NGS) technology was used to sequence the genomes of:
a) an L5178Y cell line frequently used in MLA studies.
b) Tk-/- mutant colonies induced by two prototypical mutagens in the assay.
• The goal of this pilot study was to exploit NGS technology with a view to shedding new light on the genome of cells used in the MLA, as well as the changes induced as a result of the exposure to mutagens.




