Metabolomics, She Wrote: Towards Accurate Time of Death Analysis by GC-MS

Complete the form below to unlock access to ALL audio articles.
Estimating the time of death is one of the most important and challenging tasks in a forensic investigation. As the first step towards understanding the circumstances surrounding an individual’s final moments, establishing the post-mortem interval (PMI) is an essential part of determining where, how, and potentially why a death has occurred.
Pathologists can estimate the time of death with some confidence up to 24 hours after death, based on visual inspection of the body and other assessments. However, this accuracy depends on the experience of the investigator, as well as the environmental conditions to which the body has been exposed. Once the cadaver has equilibrated to ambient temperature, establishing PMI becomes much more challenging. As a result, a laboratory method based on a robust biomarker for PMI would help to improve the reliability of time of death analysis.
The promise of metabolomics
Metabolomics may offer a more reliable approach to PMI determination. Metabolomics involves the characterization and quantitation of the small molecules present in biological systems. These include amino acids, sugars and phosphosugars, as well as biogenic amines and lipids. As the body starts to decay post-mortem, changes in the levels of these compounds can help to pinpoint the length of time since death.
Untargeted metabolomics can be challenging due to the need to identify and quantify hundreds of different unknown compounds. Gas chromatography-mass spectrometry (GC-MS) is commonly used for metabolomics applications as it provides exceptional chromatographic resolution, reproducibility and peak capacity. However, the technique has been limited by a lack of mass spectrometry support. Using conventional GC-MS technologies, achieving the necessary mass resolution, dynamic range and scan rate to confidently analyze complex biological samples has proven difficult.
Recent advances in metabolomics analysis
The GC-MS landscape is now changing, with the advent of new technologies that are specifically designed to overcome existing metabolomics challenges. For example, systems based on high resolution accurate mass (HRAM) instruments are driving unprecedented levels of mass resolution and achieving sub-ppm accuracy, giving researchers greater confidence in the data they collect.
With key metabolites present at a wide range of levels in biological samples, the analytical techniques employed to profile the metabolome must be capable of generating reliable results at both high and very low analyte concentrations. The latest GC-MS systems offer a wide dynamic range that is capable of delivering reliable results, even with complex samples. Moreover, with untargeted metabolomics pipelines reliant on consistent data deconvolution to detect species from overlapping total ion chromatogram peaks, the high scan speed offered by the latest instruments is helping to accelerate the delivery of critical metabolomic insight.
Determining post-mortem interval using GC-MS
The power of modern GC-MS instruments is unlocking new opportunities for PMI analysis. Recent findings highlight how these latest technologies, when used alongside statistical analysis tools, could enable the adoption of a metabolomics-based approach to time of death analysis.
Rat muscle tissue samples decayed over a period of 0–3 days at room temperature were analyzed using a GC-MS system. Following deconvolution, distinct electron ionization (EI) peak clusters were identified and quantified across the dataset and investigated using univariate and multivariate statistical analysis (Figure 1).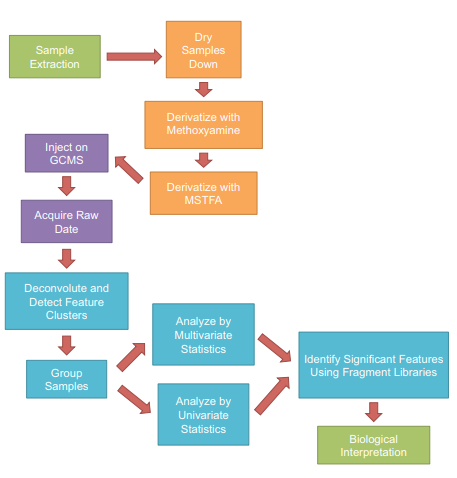
Figure 1. Workflow used for the metabolomics study. Color-coding shows work package assignment: green for wet lab biologists, orange for lab technologist, purple for instrumentation, and blue for informatician.
The partial least squares discriminant analysis plot (Figure 2) condenses the large number of metabolites to principal components that encompass the majority of variance in the dataset. The resulting plot shows that the tissue samples taken immediately post-mortem (T0 shown in green) cluster together and differ significantly from the decomposing tissue samples (T1, T2 and T3, shown in blue, red and yellow respectively). These results highlight how metabolomics can be an effective tool to provide information on PMI.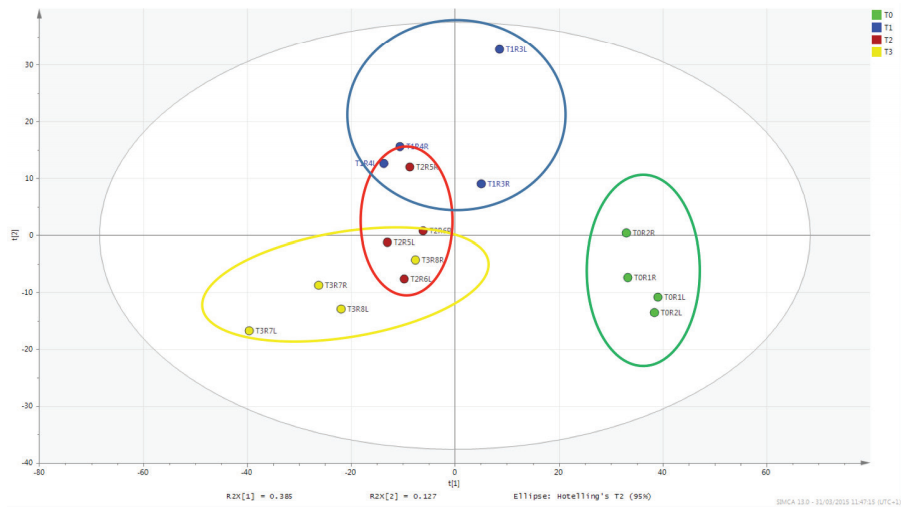
Figure 2. Partial least squares discriminant analysis model of the rat muscle tissue decomposition study.
Furthermore, the EI data could be compared against the National Institute of Standards and Technology (NIST) commercial library for tentative compound identification. The most significant metabolomic changes could be matched with high confidence to amino acids and other compounds associated with decomposition, and were further supported by spectral matches with mass fragments.
Towards a metabolomics approach to time of death analysis
In the search for reliable answers in PMI analysis, advances in GC-MS systems are opening up new opportunities for forensic investigations. The ultra-high resolution and consistent sub-ppm accurate mass measurements that can be achieved using the latest instruments are driving more confident metabolomics analyses. Moreover, thanks to the wide dynamic range of these new systems, and ease with which tentative compound identification can be performed, a metabolomics approach to establishing PMI could be just around the corner.

