Designing Vaccines Using Artificial Proteins
Complete the form below to unlock access to ALL audio articles.
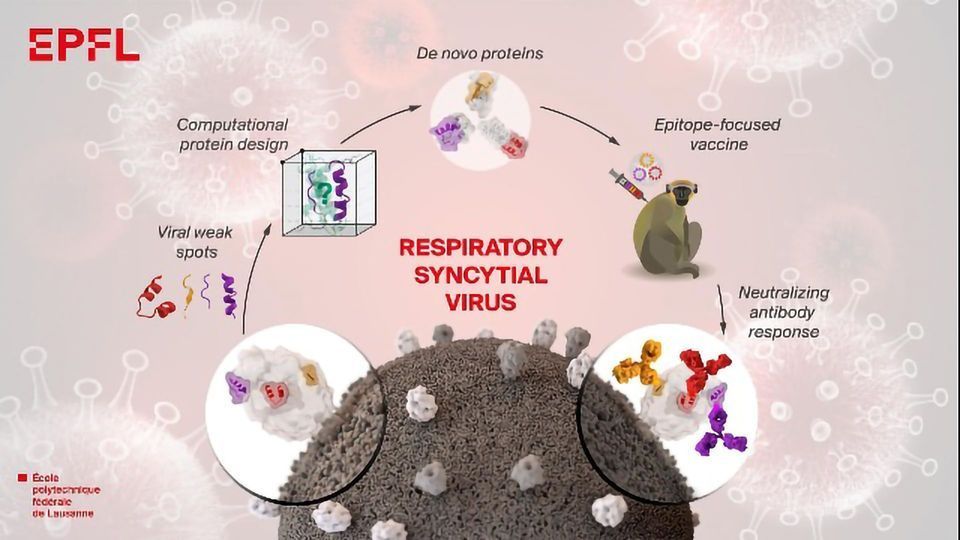
Vaccines are one of the most effective interventions to prevent the spreading of infectious diseases. They trigger the immune system to produce antibodies that protect us against infection. However, we still lack efficacious vaccines for many important pathogens, like the flu or dengue fever. "When a vaccine doesn't work well, we tend to think that it's because the antibodies produced are not protective," says Bruno Correia, a professor at the Laboratory of Protein Design & Immunoengineering (LPDI) in EPFL's School of Engineering. "It's usually because our immune system is simply making the wrong type of antibodies". Scientists in Correia's lab have now developed a strategy to design artificial proteins that very precisely instruct the body's immune system which antibodies to produce. The study has been published in the journal Science.
Building proteins like Legos
The EPFL team created artificial proteins designed using computational methods. "They don't exist in nature," says Che Yang, a PhD student and co-leading author in the study.
"We developed a protein design algorithm called TopoBuilder. It lets you construct proteins virtually as if you were putting Lego bricks together. Assembling artificial proteins that have novel functions is absolutely fascinating." says Fabian Sesterhenn, a PhD student and co-leading author.
A disease without a vaccine
Correia's team focused on the design of de novo proteins that can result in a vaccine for the respiratory syncytial virus (RSV). RSV causes serious lung infections and is a leading cause of hospitalization in infants and the elderly, "Despite several decades of research, up to today there is still no vaccine or cure for respiratory syncytial virus," says Correia.
The artificial proteins were created in the laboratory and then tested in animal models, and triggered the immune system to produce specific antibodies against weak spots in RSV. "Our findings are encouraging because they indicate that one day we will be able to design vaccines that target specific viruses more effectively, by prompting the immune system to generate those particular antibodies," says Correia. "We still have a lot of work ahead to make the vaccine we developed more effective - this study is a first step in that direction."
Methods for creating de novo proteins have applications well beyond immunology - they can also be used in various branches of biotechnology to expand the structural and functional range of natural proteins. "We can now use the protein design tools to create proteins for other biomedical applications such as protein-based drugs or functionalized biomaterials," concludes Sesterhenn.
Reference: F. Sesterhenn, C. Yang, J. Bonet, J. T. Cramer, X. Wen, Y. Wang, C. Chiang, L. A. Abriata, I. Kucharska, G. Castoro, S. S. Vollers, M. Galloux, E. Dheilly, S. Rosset, P. Corthésy, S. Georgeon, M. Villard, C. A. Richard, D. Descamps, T. Delgado, E. Oricchio, M. Rameix-Welti, V. Más, S. Ervin, J. F. Eléouët, S. Riffault, J. T. Bates, J. P. Julien, Y. Li, T. Jardetzky, T. Krey & B. E. Correia. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science
This article has been republished from the following materials. Note: material may have been edited for length and content. For further information, please contact the cited source.

