Viral Comparisons

Complete the form below to unlock access to ALL audio articles.
Viruses are tiny—merely millionths of a millimeter in diameter—but what they lack in size, they make up in quantity.
“If you were to take all the viruses from the planet, and lay them side by side, the length would be 1,000 times the length of the Milky Way,” said David Ussery, who leads the comparative genomics group at the Department of Energy’s Oak Ridge National Laboratory.
This universe of viruses is largely unexplored, even as new viruses are regularly identified in metagenomic studies that sample and sequence different viral species. Scientists use advanced genetic sequencing methods to sequence hundreds of viral genomes in a matter of hours. But making sense of the data is a challenge.
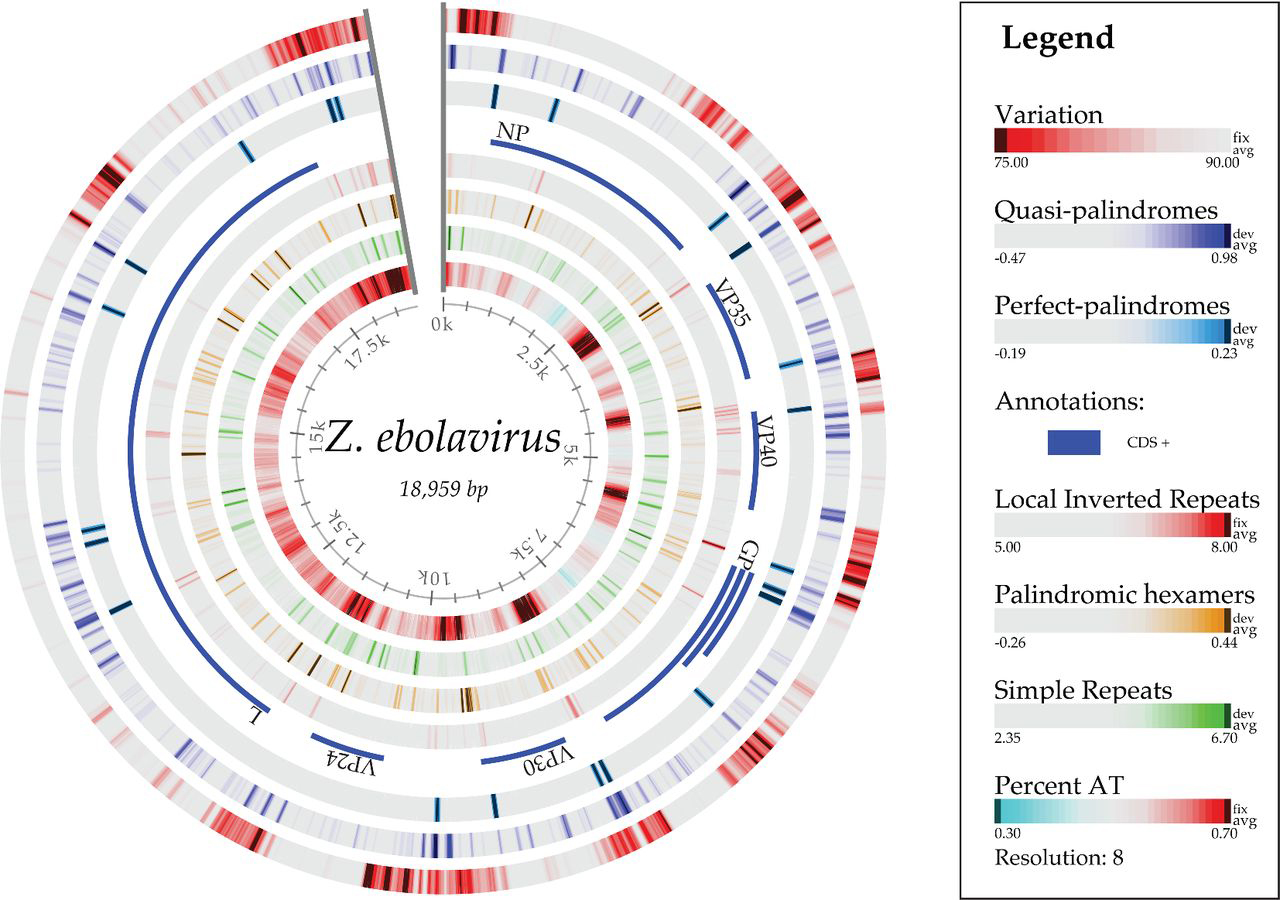
An ORNL team of comparative genomics and computational science researchers is attempting to bring some order to this viral data overload. In a recent study Ussery and a team of researchers compared approximately 4,000 complete virus genomes downloaded from a public database known as GenBank.
By compressing the sequence files, the team created a virus dendrogram that maps out the relationships among all the different virus families. Ussery notes the figure is not a true evolutionary tree. Rather, it roughly approximates how divergent sequences are from one another.
“No one has published a dendrogram like this of all viruses,” he said. “This is an example of where we want to go in terms of building up the computational facilities to do these kinds of comparisons.”
The team’s ongoing biosciences and computing collaboration will benefit other ORNL research projects that rely on comparative genomics and environmental metagenomics. The storage infrastructure for viral genomes will serve as a template and prototype for bacterial, fungal and plant genomes, which are used in the lab’s Plant-Microbe Interfaces and BioEnergy Science Center projects.
“Viral genomes are short – one million times smaller than human genomes,” Ussery said. “In principle, it should be million times easier. You have to start with something small and tractable and build your way up.”
As an example, the researchers used the comparative map to more closely analyze sequences in the Filovirus family, which includes the Ebola and Marburg viruses. Comparing genomes and understanding variability within an individual virus’s genome could inform efforts in preventing or responding to outbreaks such as the Ebola epidemic in West Africa. Ussery explains it would be possible to take the genome sequence for a new virus and quickly find the nearest neighbors.
“The idea in the future, if there is an outbreak, you can use the genome sequences collected from individuals across the country to track the spread in real time, in much the same way that we track the weather,” he said.
Likewise, comparative genomics is helpful in directing immunologists toward potential treatments. Collaborators on the paper, led by Ole Lund from the Technical University of Denmark, used the comparative sequencing analysis to look for specific regions of an Ebola virus protein that could be potential targets for vaccine development.
Enabling collaboration among genomics, computational and health experts is a key role for ORNL, explains Michael Leuze, who leads the lab’s Computational Biomolecular Modeling and Bioinformatics group. He and Ussery envision establishing a center that leverages lab expertise in genomics, computer science and big data and neutron science.
“Many of the technologies and techniques that we use on regular basis are applicable to viruses,” he said. “Dealing with viral outbreaks is outside of our scope, but understanding the genomics of viruses is an area where we can make a real contribution.”

