Optimizing Kinetics in Drug Discovery

Complete the form below to unlock access to ALL audio articles.
As biology presents ever-more challenging targets, the chemistry world is pushing ahead with new tools to design or discover drugs that have optimal kinetics – they bind well to their target, exert the desired effect and have acceptable toxicity profiles. We look at two different approaches being used to explore and optimize the kinetics of tomorrow’s medicines.
The medicinal chemist’s approach
Paul Brennan is a medicinal chemist working on designing chemical probes that can interrogate novel biological targets and design drugs with the right kinetics to exploit them.
The approach has shown a great deal of success in revealing biological insights which can be further investigated to determine their therapeutic impact. For example, in 2014 they were able to identify small molecule ligands for the bromodomain of the CREB binding protein (CBP)/p300 lysine acetyl transferases that are key transcriptional coactivators in human cells.1
“CBP and p300 are involved in many cell processes, but it wasn’t known whether targeting the bromodomain would affect these functions,” explains Brennan. “So we designed a chemical probe for the bromodomain which we then shared with collaborators.” A key discovery soon followed. “It turned out that if you inhibit the bromodomain of CRB/p300 it affects interleukin-17 signaling, which is a key inflammatory molecule involved in autoimmune diseases such as ankylosing spondylitis and psoriatic arthritis.” 2 The inhibitor is now being tested in a debilitating fibrotic disorder called Dupuytren's disease, as a topical treatment instead of or following surgery.3
Now Brennan’s team is turning its attention to a less understood area – epitranscriptomics, the study of RNA modifications. “RNA is not just a string of nucleic acids, it has modifications to it that change its base pairs, and it has caps put on the ends of it to determine whether it will ultimately be turned into protein. There are enzymes that catalyze those modifications, but it’s a fairly new field and so we want to make small molecules that can help us understand what these enzymes do.”
They are also looking at covalent compounds, which had been overlooked for several years because of concerns with potential toxicity. Brennan is collaborating with Nir London at the Weizmann Institute in Israel on screening covalent compounds. “We found lots of interesting hits and the big advantage of these being covalent compounds is that they're really potent already compared to reversible inhibitors. People worry about the toxicity of irreversible compounds but with our fragment approach we can find the least reactive covalent compounds that will be more selective and less toxic.”
“The big challenge now is to generate these chemicals much more quickly,” says Brennan. “A lot of it will come from algorithms that help us design molecules better.”
Brennan continues: “The slow and expensive step is always making the compounds and 99% of what we make isn't very good. If we can get the quality up – for 10% or even 50% of the molecules we make – it would be much faster and cheaper making chemical probes. That’s where we want to go in the future.”
Fragment Based Drug Discovery on Pioneer Systems Using Next Generation SPR Analysis
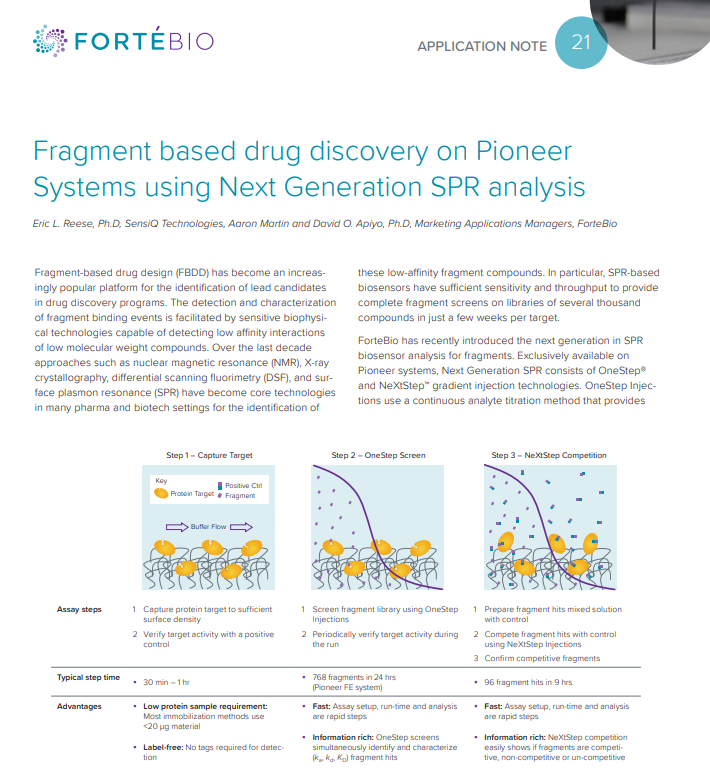
ForteBio has recently introduced the next generation in SPR biosensor analysis for fragments, exclusively available on Pioneer systems. Download this application note to learn about ForteBio's Next Generation SPR, including fragment screening with OneStep and NeXtStep Injection techniques and a comparison of OneStep and conventional SPR in fragment-based drug discovery.
Download App NoteSponsored Content
The chemical engineer’s approach
Greg Thurber is a chemical engineer who works on optimizing the delivery of macromolecules and molecular imaging agents into the body. “Our goal is to predict ahead of time using computational models how macromolecules move, such that we can design agents that are going to work more efficiently.”
The lab is using molecular imaging tools to follow these drugs across multiple different length scales, explains Thurber. “We can follow them in the whole organism via the blood, we can go down to the tissue level to see how they are diffusing across length scales equivalent to the thickness of a human hair, and right down to the cellular and subcellular level within a cell. We can figure out exactly where these drugs are going, how quickly they are getting there and whether they are being broken down in the process."
The main workhorse of the approach is near infrared fluorescence imaging which literally lights up the cells and tissues to allow tracking of the drugs using a simple chemical tag. So, what comes first? The mathematical model or the molecular imaging?
“It’s an iterative process,” says Thurber. “At this point the research is far enough advanced that we can make fairly accurate predictions just based on the structure of a molecule. For the most part we start with a computational model and predict how that drug is going to distribute within the body.” This takes into account factors like the size of the drug, how tightly it binds, its target in the body and other types of interactions the molecule has (such as whether it binds to other cells within the body) as well as other parameters like blood flow into a tissue.
“Then we would go ahead and do the experiment in vivo to really compare: are the molecules behaving as we predicted? Is the drug that was predicted to be more efficient and more effective actually reaching more cells?”
This combined approach of modeling and microscopy allows studies that were not possible a few years ago, Thurber says. “Although mathematical modeling of drug delivery is not new, we've now reached a turning point in the simulations where we can make predictions. With older models you had to collect data from in vivo experiments and then fit a line to the data points. What is transforming the field in my opinion is that now we get to take a blank sheet of paper and draw the line and say this is what's going to happen. Then we can do a very focused number of experiments to show that the data points line up with our predictions.”
One example highlighting the potential of this combined approach is in the field of antibody-drug conjugates, which comprise a monoclonal antibody attached to a very potent small molecule drug. This highly targeted class of biopharmaceutical is having something of a renaissance in oncology. “The concept behind these drugs is very simple, but in practice their delivery to tumors is very inefficient,” explains Thurber. “Our models have enabled us to predict exactly how the properties of these drugs impact their distribution, and we’ve been able to show that you can manipulate the dosing and the structure of these drugs to improve efficacy.4
A second example is for optimizing drugs that are designed to interfere with targets located inside of cells. “Typically biologics cannot reach them [intracellular targets], as they're too big to get inside of the cell. If you put a very high positive charge on these molecules they can readily get inside cells, but if you try and deliver this intravenously it's going to be rapidly cleared by the liver and other tissues. Computational models can help you balance the properties to make sure the drug would reach its target in vivo.”5
Thurber’s ultimate goal is to be able to take the structure of a drug and know exactly how it's going to distribute in the body from the whole person to the different organs, tissues, cells and subcellular components. “There are a few parameters that we still need to get a better handle on such as membrane permeability, but I think that's an achievable and reachable goal within the next decade,” says Thurber.
A Guide to Measuring Drug-Target Residence Times with Biochemical Assays

During drug development initiatives, analysis of drug-target residence times can improve efficacy, increase therapeutic window, and reduce the risk of premature focus on candidate compounds that are likely to have undesirable side effects. Download this guide to learn about how residence time assays provide insight into drug-target interactions and the basics of transcreener assays.
Download GuideSponsored Content
The complex kinetics of today’s drugs and targets
Whereas traditional small molecule drug discovery has had well established rules (e.g. Lipinski’s Rule of Five),6 with undruggable targets and newer types of drugs the rules are changing. The two distinct approaches discussed above suggest that computational technologies and algorithms will be at the heart of the new era of studying drug kinetics.
References
1. Hay DA., et al. Discovery and Optimization of Small-Molecule Ligands for the CBP/ p300 Bromodomains. J. Am. Chem. Soc. 2014; 136: 9308−9319
2. Hammitzsch A., et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. PNAS. 2015; 112: 10768−10773
3. Williams L., et al. Localised inhibition of histone acetylation may offer a novel therapeutic strategy to down regulate the myofibroblast phenotype in Dupuytren’s disease. Drug Discovery 2016 Conference, Poster number 121 [online] Available: https://www.myeventflo.com/event-lecture.asp?lectID=11426
4. Cilliers C., et al. Improved tumor penetration and single-cell targeting of antibody-drug conjugates increases anticancer efficacy and host survival. Cancer Research. 2018; 78: 758−768.
5. Atangcho L., et al. Hitting Undruggable Targets: Viewing Stabilized Peptide Development through the Lens of Quantitative Systems Pharmacology. Trends Biochem Sci. 2019; 44: 241−257
6. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004; 1: 337−341


