Averting the Antibiotic Resistance Apocalypse

Complete the form below to unlock access to ALL audio articles.
The “Antibiotic Apocalypse” scenario, where drug-resistant bacteria end humanity, often seems to be somewhat inevitable, a process driven so strongly by the vast powers of industries such as agriculture and healthcare that humanity might as well lie down and let a tide of invincible bacteria rise off kitchen surfaces to smother us all.
Back in pragmatic reality, scientists are coming up with ever-more ingenious ways to combat the growing threat, exemplified by a recent paper from researchers at the European Molecular Biology Laboratory (EMBL). The authors have determined the molecular origin of a mechanism bacteria use to transfer resistance genes that fuels the rapid spread of resistance. The researchers then went on to develop compounds which can block this transfer.
Putting a Lid on Jumping DNA
Transposons are pieces of genetic code that can easily move around the genome, and, in bacteria, between individual organisms. When these pieces of code contain genes which bestow drug resistance, the recipient bacterium also becomes resistant. This means resistance can spread rapidly around a population of bacteria and explains how bacteria such as MRSA (methicillin-resistant Staphylococcus aureus) and VRE (vancomycin-resistant enterococcus) have gained resistance to our best-available antibiotics so quickly.
The Barabas group at EMBL focuses on these transposons, also called jumping DNA. The group’s work has revealed the crystal structure of a molecular machine, fashioned out of DNA and proteins, that inserts transposons into bacteria, facilitating the spread of resistance (below).
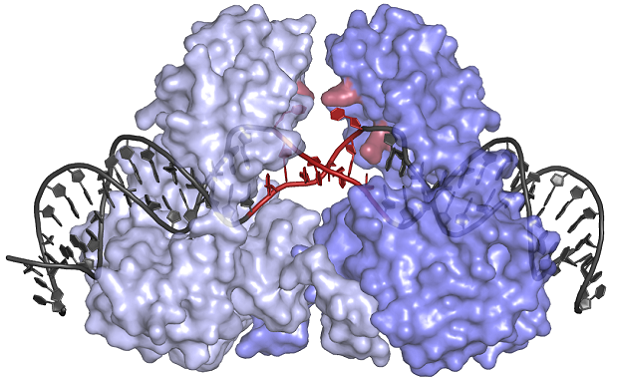
The unusual shape of the protein component of the machine protects the transposon DNA that it shuttles from damage, and can force open the transposon, which means it can be easily seeded into genomes at multiple locations.
Senior author and research leader Orsolya Barabas explained how the protein facilitates the spread of genes: “If you think of ropes or wires, they are usually bundled and wound-up to make them stronger. If you want to tear or cut one, it’s much easier if you unwind and loosen it first,” says Barabas, “It’s the same for DNA, and the transposon transfer mechanism takes advantage of this.”
Detailing the crystal structure of this protein opened the gate to developing a strategy to prevent the transfer mechanisms, and Barabas’s team leapt at the chance, creating not one but two ways of blocking the transposons.
The first prevents the protein, called integrase, from activating and inserting its cargo by physically restricting its movement with a specially designed peptide chain. The second, more cunning, method uses a DNA mimic, which interferes with the protein’s attempt to integrate the DNA into its target gene. A common problem with such techniques is the risk of accidentally targeting other gene elements in benign bacteria, but Barabas plays down that risk. “As we believe these features are broadly present in these jumping DNA elements, but not in related cellular systems, they may be quite specific to transposons. This way, we can target only the bacteria we want, and not the many good bacteria in our bodies and the environment.”
Tackling the Causes of Antibiotic Resistance
The potential for this research is huge, as Barabas explains: “Our discoveries will allow us to design improved diagnostic and preventive approaches, so as to better control the spread of antibiotic resistance genes. In addition, these insights can allow us to discover strategies to block resistance transfer, and our current study already provides proof-of-principle for this. Such strategies could, for example, help prevent resistance transfer in people diagnosed as carriers of antibiotic resistant bacteria.”“However, there is still a long way to go from a molecular structure to such applications. Therefore, our lab now focuses on better understanding the transfer mechanisms in vitro in microbial communities and in real life, and on testing and further developing strategies to limit transfer. We collaborate with research groups internationally to study these both in the lab and in the clinic.”
These promising collaborations with other research groups show that science isn’t taking the end-of-days warnings about antibiotic resistance lying down, but Barabas is clear that research cannot win this war alone.
“Research helps better understand the modes and requirements of drug resistance development and spreading. These insights will allow us to develop improved diagnostic and risk assessment approaches, and to design improved treatment procedures for antibiotic resistance colonized patients to prevent further transmission of resistance. However, broad public awareness to the threat of antibiotic resistance spreading and changes in agricultural and healthcare procedures are also very important if we want to win this battle. The overuse of antibiotics in medicine and farming is an important accelerator of multi-drug resistance development.”
Reference: Rubio-Cosials, A., Schulz, E. C., Lambertsen, L., Smyshlyaev, G., Rojas-Cordova, C., Forslund, K., … Barabas, O. (2018). Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance. Cell, 0(0). https://doi.org/10.1016/j.cell.2018.02.032


