Practical Guidance for the Confident Application of Differential Scanning Calorimetry
Complete the form below to unlock access to ALL audio articles.
The challenges of achieving reproducibility in life science research are well-documented1, and there is widespread recognition that improvements in this area are essential to provide a more secure platform for ongoing progress. A recent report from a group of researchers at Bayer underlined the scale of the problem, finding that over 75% of published data on potential drug targets could not be replicated2. Poor reproducibility can be attributed to a number of factors, but one important area for improvement is greater awareness of the analytical techniques which offer the greatest insight into a critical problem, and how best to apply them.
In this article, we provide practical guidance for the application of differential scanning calorimetry (DSC), a core technique for studying the stability and higher order structure (HOS) of proteins. We explain how DSC works, and highlight good practice for ensuring optimal data quality. The interpretation and value of the resulting data is also discussed.
DSC: the basics
DSC systems measure changes in the molar heat capacity of a sample, which are triggered by the application of heat. Figure 1 shows the thermal core of a DSC system, which incorporates cells for reference and sample solutions.
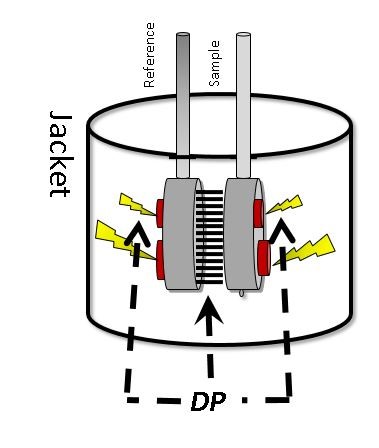
Figure 1: Schematic of the thermal core of a DSC system showing the reference and sample cells.
During analysis, energy is applied to both cells to induce an identical, closely regulated temperature change. In a protein sample, this heating triggers unfolding, a process of conformational change that absorbs heat and at the same time alters the heat capacity of the solution (see Figure 2). The unfolding or denaturing process creates a disparity between the amounts of energy needed to maintain the temperature of the sample and reference (no protein) cells. The thermodynamic profile of this conformational change is created from the measurement of the excess energy requirement of the sample cell.

Figure 2: DSC data is presented in the form of a thermogram, a plot of specific heat capacity against temperature, with protein unfolding initially associated with a rise in specific heat capacity.
For proteins, the primary metrics derived from DSC data are:
• Thermal transition midpoint (melting transition midpoint or melting point) (TM). For a protein that reversibly denatures, TM is the temperature at which the populations of native and denatured protein are equal. A higher TM is therefore indicative of greater protein stability.
• Enthalpy of denaturation (ΔH). The enthalpy change measured during DSC experiments is the energy associated with disruption of the interactions that stabilize the HOS of the protein. As a result, DSC data can form a ‘fingerprint’ for the direct comparison of structural conformation in comparability studies and biosimilar development.
Other metrics associated with the shape of the curve, such as Tonset, the temperature at which denaturation begins, can also prove useful when assessing protein stability. This ability to efficiently generate multiple metrics relating to stability makes DSC valuable all the way from early screening, where it can be used to robustly identify potential candidates that are likely to present development challenges, through to manufacturing, for batch-to-batch comparability testing.
Optimizing data quality
With regard to DSC data quality, there are three main areas where good practice can make a substantial difference: sample preparation; instrument cleaning/performance checking; and in the selection of run conditions. While good practice can be achieved via a combination of rigorous training, effective method development and skilled operation, features of the latest automated DSC systems can substantially ease the analytical burden, at the same time boosting productivity, as highlighted below.
Sample preparation
Sample dialysis against the buffer/reference solution is a valuable preparatory step that ensures that the reference and sample solutions are identical, except for the presence of the protein. An alternative approach is to use elution buffer collected during the final step of the protein purification procedure as a reference. Both techniques enhance the ability of the instrument to precisely detect changes in the specific heat capacity associated with the protein, which are small relative to the heat capacity of the solvent, thereby maximizing sensitivity.
An additional, essential element of sample preparation is sample degassing to make sure that unfolding occurs under constant volume conditions, an underlying assumption of subsequent data analysis.
It is important to determine protein concentration using a suitable method, such as UV-visible spectroscopy. The molar concentration of the protein solution is required for full data analysis of the DSC thermogram, and concentration is also needed to determine the enthalpy and heat capacity changes associated with thermal denaturation. DSC-measured parameters such as TM can be concentration-dependent, so analysis should be performed using a range of protein concentrations. Finally, when DSC is used for stability screening, the same protein concentration should be used for each sample.
Instrument cleaning/performance checking
Rigorous cell cleaning, using water and/or detergent, prevents cross-contamination and is a prerequisite for sensitive measurement. A buffer-buffer scan is a simple but highly effective way of confirming no measurable difference in the heat input to each cell, indicating a clean system with no contaminating sample carryover, operating correctly. Performance checking against a known standard at regular intervals is also good practice when it comes to maintaining highly reproducible measurements.
In the latest DSC systems, these repetitive but critical aspects of measurement are streamlined and automated, which is especially helpful when applying DSC in regulated environments. By providing consistent, reliable cleaning, newer DSC systems reduce the need for buffer-buffer scans and performance checking, thereby maximizing sample throughput, while at the same time enhancing data quality (see Figure 3).

Figure 3: Enhanced cleaning protocols reduce the need for baseline scans, boosting sample throughput. The sixteen thermograms of ribonuclease A shown here were produced in 20 hours using an automated system, with excellent reproducibility.
Selecting run conditions
When it comes to running the DSC experiment, scan rate and temperature range are the primary parameters requiring careful consideration.
Scan rate is the rate at which the temperature of the cells is increased, with higher rates equating to faster analysis. However, the scan rate applied can influence the results obtained. For example, higher scan rates have been linked with broadening of the thermal transition peak in certain proteins3. Measuring at different scan rates during method development to determine any effect in a newly-tested sample is prudent.
The selected temperature range must be sufficiently broad to capture the complete thermal transition and allow the accurate calculation of ΔH, although an incomplete thermogram may be adequate for the detection of TM.
Again, modern systems with advanced software offer supportive features in this area, such as sample templates for the easy design, storage and duplication of identified methods. Such features ease the programming of DSC experiments, help to eliminate user-to-user variability and, as with automated performance checking, are especially helpful for ensuring the rigor required for operation within a regulated environment.
Maximizing informational output
The raw output data from a DSC experiment is a plot of heat input rate as a function of temperature. Processing this data to generate a thermogram and extract the metrics of interest involves:
• subtraction of the buffer-buffer baseline
• creation of an integration baseline
• curve fitting.
This data processing can be arduous, typically involving multiple data files and an element of human decision-making, in which case consistency can also be an issue.
Advances in DSC software directly address this challenge, accelerating data analysis by enabling the simultaneous processing of multiple datasets and making it easier to fully exploit the informational value of high sensitivity DSC measurements. For example, with the latest smart software, users can simultaneously apply the same fitting method and integration baseline to multiple thermograms, to minimize subjectivity and remove inconsistency in the data analysis process. Furthermore, an established fitting method can be saved for application to other thermograms in the study, to guarantee consistent fitting methods for all samples, by all users. This improves the reliability of TM and ΔH calculations.
The other area of data analysis where human subjectivity is usefully eliminated is in thermogram comparison. Where DSC is being used alongside other assays to confirm that a manufactured protein is ‘highly similar’ to a reference protein, to assess the effect of changes to the manufacturing process, for instance, or in the development of a biosimilar, it is crucial to detect any differences between thermograms. Innovative software tools are able to perform data analysis and evaluation on multiple DSC thermograms to provide an objective, quantitative similarity comparison to a reference. These tools, in combination with exemplary measurement practice and data processing, boost DSC’s value in comparability studies and biosimilar development.
In conclusion
The optimal application of an analytical technique relies on understanding how to develop a robust method to optimize the quality of the raw data, and how to process that data to exploit the informational value within it. DSC is a powerful, cost-effective technique for studying the conformational stability of proteins, well-suited to many different proteins and applicable over a wide temperature range. The guidance provided here summarizes certain steps that can be taken to enhance its application, and highlights the value of modern automated systems with features that boost sensitivity, productivity and reproducibility. Such systems enhance the ability of DSC to reliably differentiate the most inherently stable drug candidates, and the most stable formulations and/or manufactured batches, bringing value across the biopharmaceutical development cycle.
References:
2 http://www.nature.com/nrd/journal/v10/n9/full/nrd3439-c1.html?foxtrotcallback=true
3 Duruwoju, K.M. et al ‘Differential Scanning Calorimetry – A Method for Assessing the Thermal Stability and Conformation of Protein Antigen’ Journal of Visualized Experiments (JoVE), Issue 121, e55262, doi:10.3791/55262 (2017)

