Overcoming Challenges in Single Cell Analysis

Complete the form below to unlock access to ALL audio articles.
High throughput single cell RNA sequencing has emerged as a powerful tool for profiling complex and heterogeneous cell populations and dynamics. However, the lack of information on protein expression can make identifying cell types that have conventionally been defined by cell surface markers challenging, as mRNA and protein expression are often not tightly correlated.
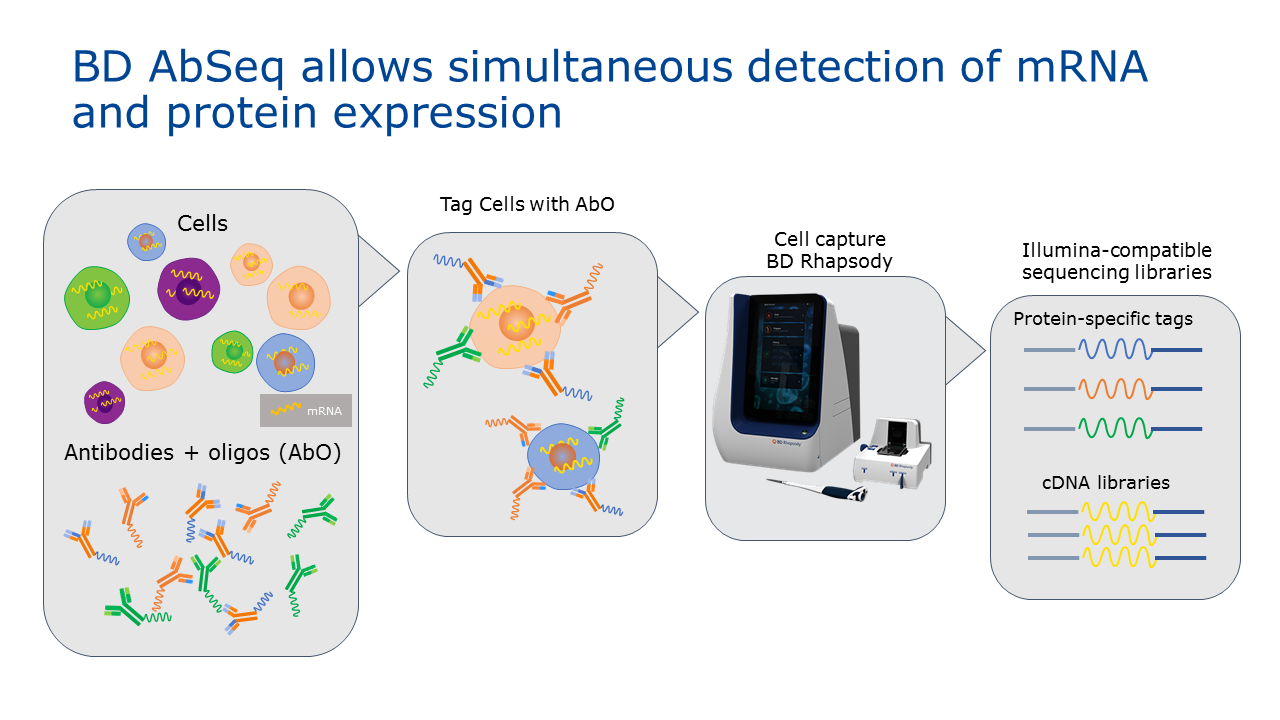
A team at BD Biosciences recently published work which demonstrates a novel approach using oligo-conjugated antibodies to measure protein expression by sequencing, which enables simultaneous detection of protein and mRNA expression in a single cell. The antibody specific oligos are captured, amplified and sequenced alongside mRNA in a single workflow using BD™ Rhapsody, a massively parallel single cell analysis system.
They believe the method they have developed has the potential to transform both single cell transcriptional profiling and high parameter proteomics and further efforts in elucidating complex biological systems, understanding disease states and enabling more effective biomarker discovery.
To find out more about the BD™ Rhapsody platform and the novel method BD have developed we spoke to Stephen Gunstream, VP Strategy, Business Development & Innovation at BD Biosciences and Christina Chang, Senior Scientist, BD Biosciences.
JR: What are the limitations of current high-throughput single cell RNA sequencing methods when trying to identify different cell types?
SG: High-throughput single cell RNA sequencing is a powerful tool to profile heterogeneous cell populations. RNA-Seq - a method of digital quantitation of individual mRNA transcripts in individual cells via technologies like molecular indexing - has begun to address cellular characterization limitation. This method creates greater diversity of labels than FACS methods and allows for the parallel processing and analysis of thousands of different transcripts for thousands of individual cells through the use of combinatorial libraries of oligonucleotide barcodes combined with common molecular biology techniques such as reverse transcription and PCR. A sequencer is then used to sequence genes, identify transcripts and produce a readout of expression levels of RNAs.
There is a need in the field to simultaneously measure two or more modalities in single cells - most specifically, transcriptome and proteome - to better understand critical differences in cells. While RNA-Seq has become a dominant technique for initial experiments for cell characterization, expression alone may not provide enough information to resolve certain biological questions. In some cases, there is not a direct relationship between the amount of RNA in a cell and the protein levels. Multiple dimensions of data improve the robustness of conclusive statements about defining cell types.
The BD Rhapsody system is poised to be the first commercial system to allow for the simultaneous quantitation of both protein and RNA within individual cells. Using extensive catalogues of high quality antibodies generated for FACS analysis and combining this with the diversity of labels achievable with DNA based barcodes, BD Rhapsody can provide simultaneous quantitation of RNA transcript levels and proteins markers for which there are antibodies, yielding new possibilities for single cell characterization.
JR: How does your approach involving oligo-conjugated antibodies help to overcome these limitations? Why is the BD Rhapsody™ platform well suited to this method?
CC: The power of this method lies within the oligo-conjugated antibody panel. The BD Rhapsody system enlists DNA-barcoded antibodies to measure protein expression. AbSeq antibodies are tagged with oligos that comprise of a PCR handle for targeted amplification, an antibody specific tag, and a poly T tail to enable capture onto beads. Using the BD Rhapsody system with AbSeq assays enables antibody-specific oligos to be captured, amplified, and sequenced alongside mRNAs in a single workflow. This approach has the added benefit of reducing experimentation time and increasing information received in a single run.
That said, the performance of an assay is very dependent on the quality of the antibody and conjugation used. At BD, we are able to leverage our large portfolio of validated flow cytometry antibodies along with BD’s proprietary conjugation chemistry for our AbSeq products.
JR: You recently published work on identifying T-cell subsets to demonstrate the power of this method. What did you find?
CC: Our research on T-cell subset identification demonstrates the BD Rhapsody’s protein and mRNA analysis capabilities to profile complex and heterogeneous cell populations and dynamics. With the addition of protein expression, we were able to greatly improve separation between T-cell subsets and identify rare populations such as a rare T-cell subset, stem memory T-cells.
To do this, we created an oligo-conjugated antibody panel that includes many common T-cell markers. The assay was applied to human peripheral blood mononuclear cells, captured on the BD Rhapsody system, and subsampled for targeted amplification and sequencing. Following reverse transcription, targeted amplification was performed and the cells were sequenced. The AbSeq assay is highly sensitive, as side by side comparison with flow cytometry data showed similar expression distributions and fold changes between positive and negative cell populations. Using AbSeq we were able to detect markers that are hard to measure at the mRNA level either due to low mRNA abundance or having isoforms with splice sites far from the 3’ end, such as CD45RA and CD45RO. Finally, consistent with what has been described in literature, we found that a significant number of markers in our panel had very different mRNA and protein expression profiles in single cells, which underlines the value and importance of multi-omics analysis.
JR: The potential impact of this method goes beyond identifying T-cell subsets. What are the most exciting potential applications?
SG: There are several applications for BD Rhapsody Single-Cell Analysis System and AbSeq technologies. BD Rhapsody is a complete system of reagents, instruments and software for targeted gene expression analysis of tens of thousands of individual cells. Using the BD AbSeq assays with this technology enables researchers to simultaneously examine gene expression and protein markers. This method has the potential to transform single-cell transcriptional profiling and high-parameter proteomics, enabling scientists to profile complex biological systems and better understand disease states for biomarker discovery.
The ability to interrogate both protein and RNA in single cells is key for profiling cells that have a low abundance of transcripts, don’t correlate well between protein and gene expression or exhibit highly similar transcriptional profiles. The application areas for these assays is very broad. Everything from developing a more granular understanding of the immune system to characterizing tumor samples after treatment by immunotherapy. The ability to interrogate both protein and RNA within thousands of single cells will shed light on the inherent complexity of how cells function and adapt in various environments.
Stephen Gunstream and Christina Chang were speaking to Jack Rudd, Senior Editor for Technology Networks.


