Chemical Snapshots and Molecular Maps with Mass Spec Imaging
Complete the form below to unlock access to ALL audio articles.
Mass spectrometry (MS) has long been an indispensable component in a chemist’s toolkit for both identifying and quantifying molecules within a sample. But using mass spec as an imaging technique became popular only in the last two decades.
The first use of mass spectrometry imaging (MSI, also known as imaging mass spectrometry or molecular imaging) was reported in the 1990s, when Dr. Richard Caprioli of Vanderbilt University, applied Matrix-Assisted Laser Desorption/Ionization (MALDI) to visualize peptides and proteins in rat tissue slices.1
With its ability to produce a multicolor chemical map of biomolecule distribution in tissues without antibodies, the technique quickly attracted the attention of pharmaceutical and clinical researchers. Today, MSI has established itself as a powerful analytical method in the biopharma and medical fields, performing much like a ‘high-throughput form of immunohistochemistry’, and enabling the discovery of important biomarkers and mechanisms of human disease.
Why Choose MSI?
For Prof. Christoph Borchers, Dept. of Biochemistry and Microbiology, University of Victoria, British-Columbia, Canada, MALDI-MSI is the tissue imaging technique of choice because of its higher multiplexity and specificity. He explained that the approach can provide absolute specificity resulting in fewer false positives, in two ways:
1) for molecules <1000 Da, the molecular weight (MW) and elemental composition can be accurately determined.
2) information about the structure of the molecules can be obtained from tandem mass spec, which provides high-resolution data on specific fragments of the molecules.
“If mass spectrometers with high resolution are used, hundreds to thousands of molecules can be imaged simultaneously, especially small molecules like lipids and metabolites,” said Borchers.
On the opposite side of North America, at the Public Health Research Institute (PHRI) of the New Jersey Medical School, Rutgers, in New Jersey, USA, Dr. Brendan Prideaux is busy finishing up a grant ahead of a trip to Europe. As a mass spectrometrist in the Analytical Imaging Center at PHRI, Prideaux routinely uses MSI for the in-situ analysis of drug and metabolite distributions within biological tissues.
Having just published a review2 on the subject, Prideaux told us that MSI has a big advantage over classical imaging techniques, like autoradiography, because it does not rely on modifying the drugs/biomolecules by the addition of a radioactive label. This ability to directly visualize drug distributions in tissues at cellular-level spatial detail, free of labels, makes the technique particularly amenable to the early stages of drug development.
“We can also visualize the actual molecule by MSI rather than tracing a radioactive label. We can resolve parent drugs from their metabolites, and that’s really a crucial factor in medical research,” said Prideaux.
For example, several essential drugs used to treat tuberculosis (such as pyrazinamide) are prodrugs, or inactive forms of a drug that need to be metabolized to perform its intended function. So, for researchers studying efficacy of these drugs, it’s important to determine whether the active form of the drug (the metabolite) is getting to bacterial clusters hidden in hard-to-reach areas, like necrotic lesions with poor blood supply.
MSI in Disease Research
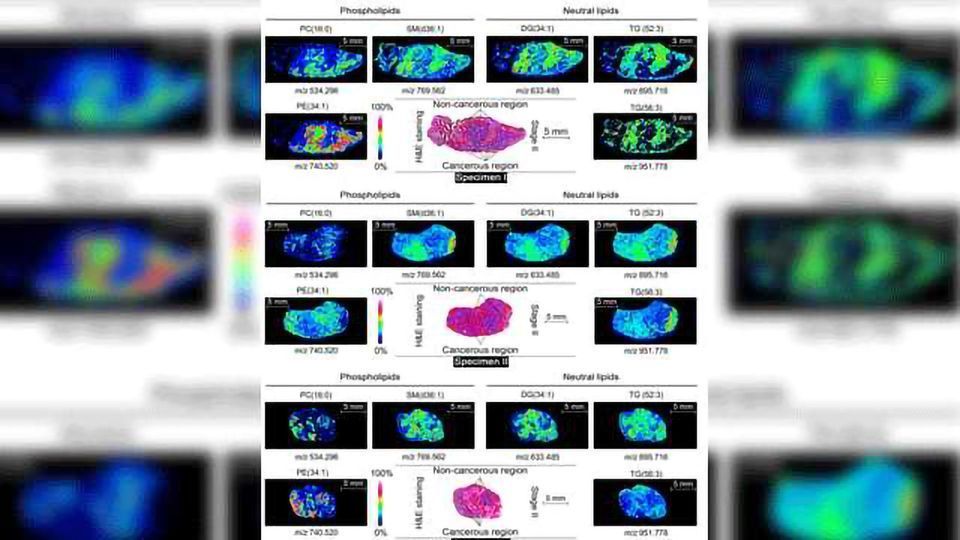
By far, the majority of applications of MSI have been in the area of cancer diagnosis, particularly for in situ detection and visualization of metabolic biomarkers associated with cancer. In one study of vulvar cancer, MALDI-MSI was used to examine three different vulvar tissue types.
Images showed differential expression of the protein cytokeratin 5 (CK5) across the tissues.3 Another study utilized MALDI-MSI to directly analyze cultured human breast cell lines and map accumulated lipid biomarkers that could potentially play a role in invasive breast cancer.4
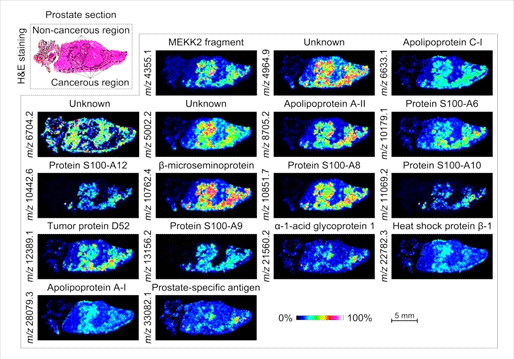
Figure 1. MALDI-MS imaging of differentially expressed proteins between the cancerous and non-cancerous regions of a human prostate cancer tissue specimen. (Image sourced from: Borchers et al. 2016, J Mass Spectrom.)
According to Borchers, one of the most significant breakthroughs enabled by MALDI-MSI was the discovery by Caprioli and his colleagues “that there are molecular changes in tumor margins that can be observed by MALDI-MSI, even though the tissue appears to be histologically normal.”
In 2010, the Caprioli group reported that these changes along tissue margins often go undetected by conventional diagnostic methods, but MALDI-MS has the specificity and sensitivity to expose them. Caprioli’s work with renal carcinoma biopsies showed how MALDI-MSI could detect changes in the tumor microenvironment. This has major implications for cancer surgery.5 In fact, the Agar group at Harvard is now using MALDI-MS to determine the boundaries of tumors.6
At the PHRI in New Jersey, Prideaux spends much of his time engaged in tuberculosis (TB) research. He has been using MALDI-MSI to monitor both TB drug distributions and markers of disease progression and/or therapeutic response. His team recently developed a MALDI-MSI method to simultaneously visualize and map out drug metabolites and bacterial lipid biomarkers within pulmonary TB lesions.7
“Using such an approach shows not only that a drug is reaching its intended target, but also that it is exerting its desired effect (or undesired effects in the case of toxicology studies),” Prideaux explained. “Our MALDI-MSI method allowed us to see where the drugs are, in relation to specific bacterial target populations. Now we are looking at applying this method to assess drug activity and other drug-induced mycobacterial metabolic responses.”
Challenges, Advances, and the Future
One of the most challenging aspects of MSI, in Prideaux’s opinion, is sample preparation. His team starts their MALDI-MSI experiments by selecting the correct combination of UV-absorbing matrix and solvent system. “Both the drug and matrix should be soluble in the solvents used and the matrix application should be sufficiently wet to extract the drug from the cells, but not so wet that it might induce delocalization and loss of spatial integrity.”
Another significant challenge lies in quantifying drug distributions. With the heterogeneous ion suppression caused by lipids and salts, the differing drug extraction efficiencies between cell/tissue types, and difficulties building standard curves for absolute quantification, it can be tough to obtain quality data.
“For our MSI studies, we normalize detected drug signals against a labeled standard drug, which we spray on the tissue surface,” remarked Prideaux. “This compensates for tissue-specific ion suppression, and we can get semi-quantitative distribution images where tissue regions with high concentrations of drug can be clearly distinguished from low concentration areas.”
In a review paper he co-authored last year, Borchers discusses the recent advances that have occurred in MALDI-MSI through the use of new matrices.8 “Conventional matrices often present problems when detecting low-MW peptides,” Borchers explained. “You see high background signals in the low-mass range and ionization efficiencies are also pretty low in the negative ion mode. But in the last few years, we’ve seen the development of many high ionization-efficiency MALDI matrices, matrix deposition procedures, and in situ sample treatments, as well as improvements of the MS instrumentation.”
Although most studies use non-targeted MSI, and provide only relative concentrations in different areas of the tissue, targeted MSI provides more sensitivity and is capable of absolute quantitation. Borcher’s review article provides insights on ways to improve the sensitivity of targeted MSI, such as on-tissue chemical derivatization. These strategies coupled with labeled internal standards enable researchers to determine the actual concentrations of these molecules per µm2 and/or the actual copy numbers per cell, which can be important measures in clinical diagnostics.
In conclusion, both Borchers and Prideaux are confident that spatial resolving capabilities of MALDI-MSI will continue to improve with advancements in laser optics and the commensurable increases in mass spectrometer sensitivity. Also, they agree that the emergence of 3D MSI for drug imaging in tissues will require sophisticated software and computational tools that can handle and process large datasets.
“With the growing potential for personalized medicine, I expect MSI to become an integral part of a multiomics approach in drug discovery and development, for both drug localization and biomarker discovery of disease pathogenesis and therapeutic response,” added Prideaux.
References:
1Caprioli, R. M.; Farmer, T. B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69 (23), 4751-4760.
2Prideaux, B.; Lenaerts, A.; Dartois, V., Imaging and spatially resolved quantification of drug distribution in tissues by mass spectrometry. Curr. Opin. Chem. Biol. 2018, 44, 93-100.
3Zhang, C.; Arentz, G.; Winderbaum, L. et al. MALDI mass spectrometry imaging reveals decreased CK5 levels in vulvar squamous cell carcinomas compared to the precursor lesion differentiated vulvar intraepithelial neoplasia. Int. J. Mol. Sci. 2016, 17.
4Wang, S.; Chen, X.; Luan, H. et al. Matrix-assisted laser desorption/ionization mass spectrometry imaging of cell cultures for the lipidomic analysis of potential lipid markers in human breast cancer invasion. Rapid Commun. Mass Spectr. 2016, 30:533-542.
5Oppenheimer, S. R.; Mi, D.; Sanders, M. E.; Caprioli, R. M., Molecular Analysis of Tumor Margins by MALDI Mass Spectrometry in Renal Carcinoma. J. Proteome Res. 2010, 9 (5), 2182-2190.
6Calligaris, D.; Feldman, D. R.; Norton, I. et al. MALDI mass spectrometry imaging analysis of pituitary adenomas for near-real-time tumor delineation. Proc Natl Acad Sci USA, 2015, 112(32), 9978–9983.
7Blanc, L.; Lenaerts, A.; Dartois, V.; Prideaux, B., Visualization of Mycobacterial Biomarkers and Tuberculosis Drugs in Infected Tissue by MALDI-MS Imaging. Anal. Chem. 2018, 90 (10), 6275-6282.
8Baker, T. C.; Han, J.; Borchers, C. H., Recent advancements in matrix-assisted laser desorption/ionization mass spectrometry imaging. Curr. Opin. Biotechnol. 2017, 43, 62-69.



