The Markram Interviews Part Two: Simulations and Experiments – How Can They Work Together?

Complete the form below to unlock access to ALL audio articles.
In the second of our exclusive three-part interview series with Blue Brain Project Founder and Director Professor Henry Markram, we discuss how brain simulations can benefit neuroscience. In silico, informatics-based approaches have revolutionized life sciences over the past three decades but using these approaches to map the brain is a challenge unlike any other in neurobiology. We hear from Markram why the challenge is one worth taking on, how in silico techniques can improve reproducibility in neuroscience and how Markram’s Blue Brain Project has made use of supercomputing resources. Find out more about our Markram Interviews series here.
Ruairi Mackenzie (RM): The brain is so complex, and we know so little, how can you even begin to build a digital copy?
Henry Markram (HM): We will need a lot more data to build and fully test digital reconstructions of the brain and there are indeed still many unsolved puzzles about how the brain works. However, it is incorrect to think of these early digital copies of the brain as something you build only when you have all the pieces of the puzzle (i.e. all the data and knowledge). We have developed this approach precisely because we don’t have, and we don’t think we can get, all the data. Our approach helps to identify and predict the missing pieces as we try to build it. The way this works is that we start by pragmatically collecting whatever data is available (e.g. on a circuit in the brain). This is very sparse data (e.g. the number of synapses between a few types of neurons) with many missing gaps (e.g. how many synapses exist between all types of neurons). We then analyze the data using data science and neuroinformatics tools to identify principles of how all the data is related to each other (e.g. when Type A neuron connects with Type B, it uses five synapses and when Type A connects with Type C, it uses on average ten synapses). We then build an algorithm that follows these biological rules. The algorithm connects and forms synapses between all the neurons according to these rules. We then test the algorithm by comparing the reconstruction with new data that we left on the side and that was not used to build the algorithm (e.g. the number of synapses between two other types of neurons). If it passes the tests, then we continue to build in more biology. If it fails, we go back and try and understand what was wrong about our understanding (e.g. of how neurons form connections) and ask whether we perhaps need more data. We measure when we need to.
Actually, it is exciting when the model fails because we know then that we are at the frontier of science, since we have considered all available data and all current theories of how neurons connect, and it still fails. That means there is something unexpected happening. Sometimes, this means going back to the lab and doing new experiments because the data may be wrong. Sometimes it means our understanding and theories about the data are wrong. So we progress our understanding each time we can falsify our model. By running through this cycle many times, we get closer and closer to the biology and increase our understanding of how the brain is designed. This allows us to make many more accurate predictions of what is missing and move on to more difficult gaps in our data and knowledge.
This approach does not replace experiments, it just accelerates finding the data and filling in our gaps in our knowledge. We follow this process strictly to build models of ion channels, neurons, synapses, circuits, brain regions and will follow it to build the first draft copy of the whole mouse brain.
RM: Why are in silico models advantageous in neuroscience?
HM: In experimental neuroscience, one has access to only a very small fraction of the biological mechanisms and in theoretical neuroscience, one abstracts the biological mechanisms; in both approaches, it is difficult to get to the detailed mechanisms underlying phenomena of the brain. In simulation neuroscience, one uses biologically faithful reconstructions of brain tissue to perform in silico (as opposed to in vivo and in vitro) experiments where all the biological mechanisms are accessible. Provided the models are close to replicas of the actual brain and that they exhibit the phenomena that one wants to study, then the detailed biological mechanisms involved in the phenomena can be identified.
But there are many other reasons.
The famous neuroscientist, Ramon y Cajal, started over 100 years ago to draw every neuron with a pencil, but today we can build the brain in digital form using supercomputers. It is actually the most systematic way to organize our data, fill vast gaps in our data, test our knowledge and make predictions of what we don’t yet know.
Let’s imagine that one could manipulate and measure every facet of the brain in the laboratory; any of the dynamic changes in the genes, molecules, neurons, synapses, brain regions etc. It would obviously be better to perform biological experiments than in silico experiments. However, we are light years away from being able to do that in the laboratory. And, even if this hypothetical scenario were ever possible, we would still want to put all this data into a faithful digital copy of the brain so we can explore it in many different ways.
I see simulation neuroscience as a way to obtain a complete map of the brain. Mapping the brain is not the same kind of challenge as mapping the human genome. Mapping the brain requires description at many different levels of the brain, all the interactions between all the elements, and how they vary with development, experience, aging, strains, species, gender and a plethora of diseases. I claim that it is impossible to, and that we do not need to, measure everything (that does not mean we should stop experiments). Simulation neuroscience offers a solution - predict most of it from inevitably limited samples of the data.
Additionally, such a model would provide a mathematical description of every facet of the brain. And only when we have been able to capture, in mathematical terms, every component, interaction and emergent phenomenon in the brain, can we truly claim that we understand the brain. Only when we can recreate the world we see and experience by running these mathematical formulations and algorithms, will we have definitive proof that we have understood.
Simulation neuroscience is also a powerful tool to test theories of the brain. In neuroscience, we have many theories about all kinds of aspects of brain structure and function, and we don’t have a systematic and standardized way to test theories. Models of the brain that are not biologically accurate can be enormously helpful for fitting data and exploring concepts but cannot claim to test a theory of the brain when the model does not contain what the brain contains.
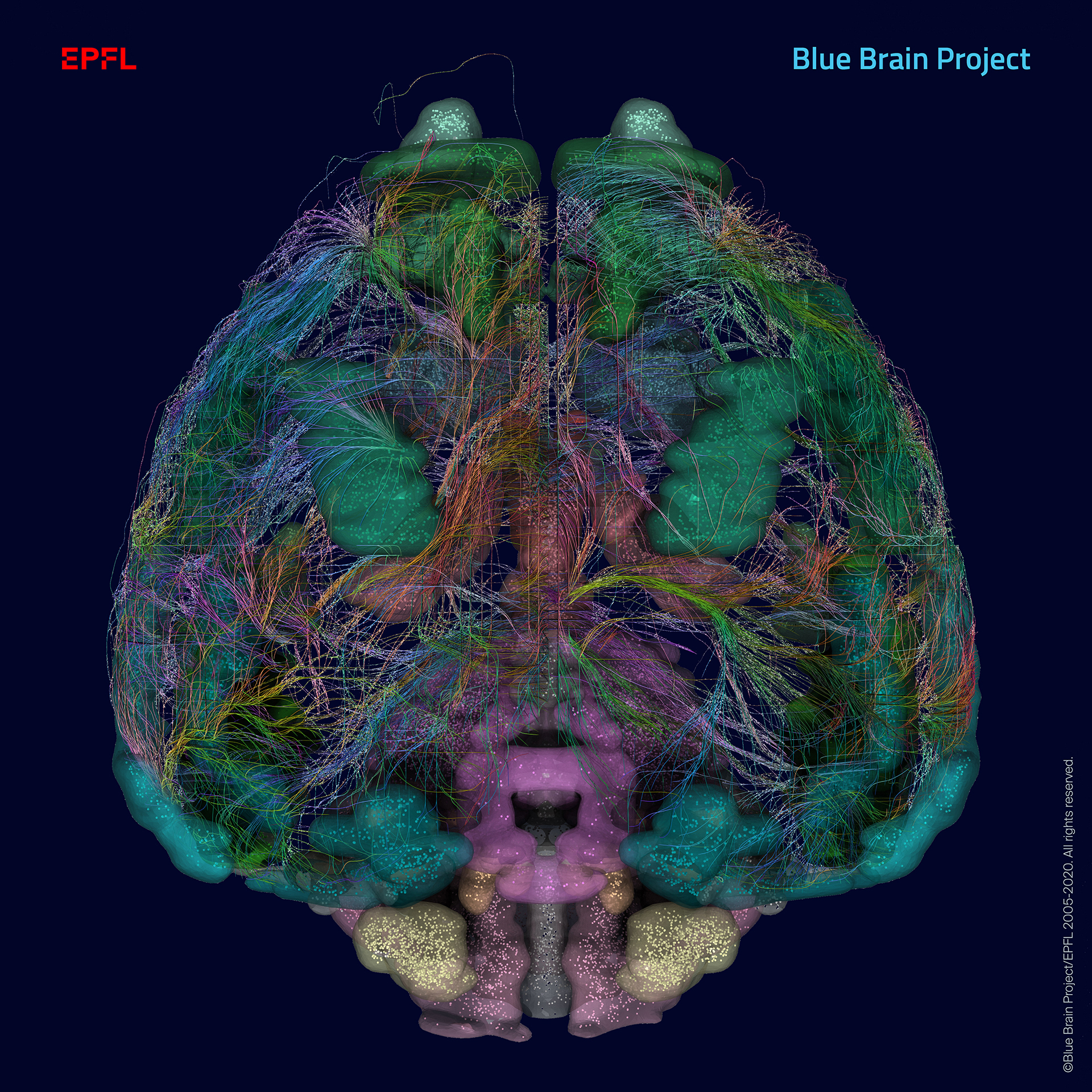
Mouse brain regions of the Blue Brain Atlas, showing individual cells (dots) and interconnections by white matter fibers (courtesy of the Allen Institute for Brain Science). Credit: Copyright © 2005-2020 Blue Brain Project/EPFL. All rights reserved.
Simulation neuroscience also brings other advantages, such as the ability to explore the sensitivity of the brain to any biological parameter, which is also a systematic path to identify all the weak points in the brain; the disease mechanisms and possible targets for intervention. Drugs target molecules, so we need models that contain molecules if we want to be able to test and design drugs in silico.
Simulation neuroscience is also important because it challenges the status quo. For example, many neuroscientists still think we first need to measure everything before we try to build a brain in a computer, even though information theory has long shown us that when there are interdependencies in the data, then one can validly predict missing data. Perhaps the most important question we need to answer is what fraction of all the data on the brain can be predicted. There is so much to measure, and experimental neuroscience is very expensive, so we should be measuring mainly those aspects that really cannot be predicted.
RM: How can in silico models improve biomedical science’s struggles with reproducibility?
HM: Neuroscience is severely hindered by a lack of reproducibility. Some studies suggest that as much as 70% of neuroscience measurements have not and/or cannot be reproduced. Since the world spends billions of dollars every year to understand the brain and its diseases, we should all be very, very worried about this problem. Today, simulation neuroscience is the only way we can think of that can systematically test for reproducibility across all reported data. The best way to test whether data is good is simply to use it; try and build with it. The flaws in the data show up quickly. In practice, we find that there are no ground truth measurements from experiments. This came as a shock to me as an experimental neuroscientist. Experimental measurements often vary far too much to be relied on as individual measurements. For example, a measurement such as the branching pattern of a particular neuron often varies more across laboratories than across species. The number of neurons in a brain region reported by different laboratories can vary more than 10 times.
One can, however, approximate the ground truth by combining different types of data. If one simultaneously considers all the reported measurements of a particular type of data (e.g. number of neurons reported in a particular brain region) and other measurements of a different type of data (e.g. number of synapses reported by another laboratory in the same brain region), then their inter-relationships constrain what is possible. The two measurements are related, since neurons form the synapses, and so if one study reports on the number of neurons it also implies how many synapses another study should find. This approach of applying multi-dimensional constraints from orthogonal datasets can become much more elaborate and be applied to many different types of data. As an experimentalist, I find it rather profound that predicted measurements can actually be more accurate than experimental measurements. As this finding becomes widely understood, it will become much more obvious why an interplay between experimental and simulation neuroscience is so important to understanding the brain.
RM: In 2018, you announced a new supercomputer, Blue Brain 5. How exactly are HPC systems like this leveraged by the Project?
HM: Blue Brain 5 is a supercomputer built by Hewlett Packard Enterprises (HPE), customized for us to tackle the many different mathematical operations we have to run when we build, simulate, visualize and analyze digital copies of brain tissue, and eventually whole brains.
Modeling an individual neuron in Blue Brain today requires solving around 20,000 ordinary differential equations. When modeling entire brain regions, we need to solve roughly 100 billion equations in each time step of the simulation (one fortieth of a millisecond).
Simulation is not the only supercomputing task we have. Digitally building the brain is a major challenge for supercomputers. For example, in a part of the brain the size of a pinhead, there are roughly a billion points where the branches of neurons touch each other. We need to find those touches and then apply biological rules to each touch point to decide if that is where a synapse can form a synapse or not based on principles of how synapses form. Most of the touch points do not form synapses, so one ends up with around 40 million touch points where synapses can form. Performing this calculation took us two weeks on our first supercomputer in 2005, but today takes seconds and we can scale this up to trillions and trillions of touch points on our latest supercomputer.
Visualization and analysis also need different kinds of supercomputing architectures. HPE is wonderful to work with; they put together a heterogeneous compute architecture that closely meets all the different workloads we have.




