Untargeted Profiling of Metabolites: In Search of Cancer Biomarkers and Antibiotics

Complete the form below to unlock access to ALL audio articles.
When Untargeted is the Way to Go
Plants and animals perform many physiological functions through the use of small molecule metabolic products. Studying these key molecular players of metabolism can tell us a lot about the phenotype of a biological system, particularly, information on disease states, dietary patterns, and drug toxicity. Metabolite profiling can be performed by targeted and untargeted methods depending on the intent of the study and the data already available. When it is necessary to accurately identify and quantify a predetermined set of metabolites (in other words, when you know what you are looking for), targeted approaches can be adopted. But when a comprehensive profile of all the metabolites in a sample, known and unknown, is required, untargeted technologies are the way to go.
Profiling Metabolites to Discover Cancer Biomarkers
One of the major research areas that benefit from metabolic profiling technologies is the identification of novel biomarkers. Metabolite concentrations change in response to various physiological processes, and observing such changes can help figure out the underlying mechanisms of the disease and the associated biomarkers. Using untargeted metabolomics, scientists can study the biofluids of diseased patients in search of these potential diagnostic biomarkers.
“Untargeted metabolic profiling is a high-throughput and massively parallel analysis in time and space,” said Professor Ching-Wan Lam, Director of Chemical Pathology, Department of Pathology, University of Hongkong, Hongkong. “You do not need to form a hypothesis (which can be wrong most of the time), and also, the chance of successfully finding a cancer biomarker is higher.”
Lam’s group employed an untargeted approach to discover new cancer biomarkers that can diagnose malignant pleural effusions. Roughly 40% of exudative pleural effusions are related to lung cancer, while ~55% are related to pulmonary tuberculosis. Since both conditions have overlapping symptoms and current diagnostic procedures are invasive and slow, a biomarker that can clearly distinguish between cancer-derived effusions and TB-derived effusions will be tremendously valuable for the clinic.
Researchers in Lam’s pathology lab use liquid chromatography-tandem mass spectrometry (LC-MS/MS) as the core analytical platform for their untargeted analyses, due to high detection sensitivity and specificity. In fact, they detected 5868 features (m/z peaks) in the positive and negative electrospray ionization (ESI) modes. Each feature has its own area-under-the-curve (AUC) values and, according to Lam, the one with the highest AUC can be distinguished as a potential cancer biomarker using further analytic tools. “LC-MS/MS is a very sensitive and unbiased technique, compared to nuclear magnetic resonance (NMR), for example,” Lam noted. “It is not possible for NMR to detect every metabolite seen by LC-MS/MS. Theoretically, one can formulate 5868 hypothesis and tested each hypothesis individually. But also, you would need 489 years in order to complete testing all these hypotheses, at a rate of one per month.”
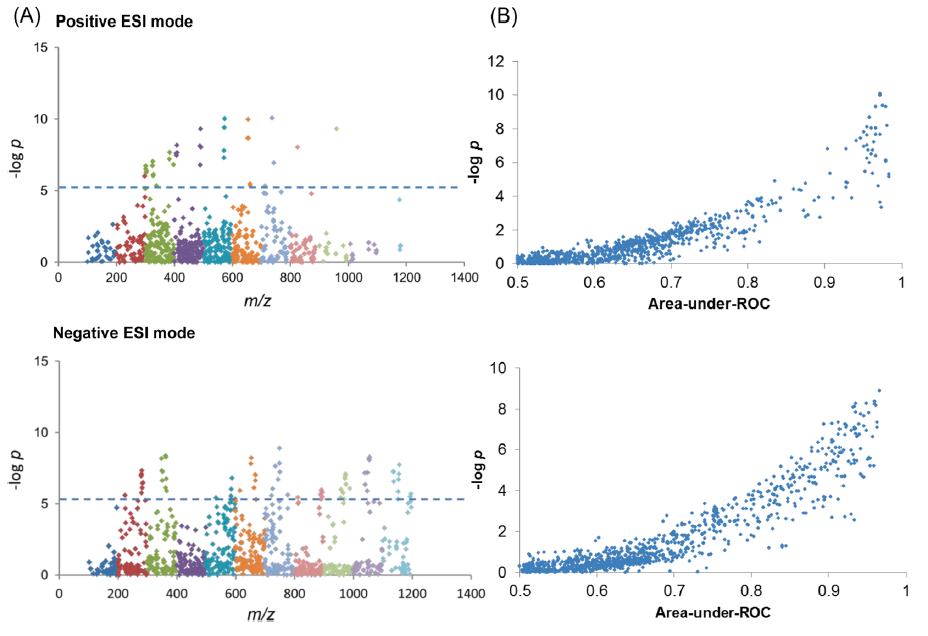
Figure 1. Data analysis in MWAS. Panel A: m/z (mass over charge ratio) of positive charge biomarkers in positive ESI (electrospray) mode and negative charge biomarkers in negative ESI mode. The dash line indicates the metabolome-wide significance level (MWSL) p-value level of 4 ×10-6. Panel B: “LASH” plot invented by Lam showing the curvilinear relationship between –log P and area-under-ROC of m/z peaks. Essentially, this is a plot of statistical significance vs clinical significance for facile identification of disease biomarkers, i.e., biomarkers with area-under-ROC >0.9 and p-value less than MWSL. (Source: Lam and Law. J Proteome Res. 2014)
Metabolome Wide Association Studies
By using Metabolome-Wide Association Studies (MWAS), Lam’s team found that most diagnostic cancer biomarkers in pleural effusions are all free fatty acids. Essentially, it is a case-and-control association study involving large-scale metabolic phenotyping of human biofluids from a disease group and a non-disease group. The MWAS workflow adapted by Lam is detailed in Figure 2 below. He emphasized that the sensitivity of his method, which yielded fatty acids as effective biomarkers, was much higher than other techniques. “There are studies which explore circulating tumor cells in pleural effusions, for instance, but the sensitivities range from 30-50%, compared to 93.8% in our case.”

Figure 2. Steps in unbiased cancer metabolic biomarker discovery using MWAS and untargeted LC-MS/MS (liquid chromatography- tandem mass spectrometry). MWAS is a case and control association study involving a large-scale metabolic phenotyping of human biofluids of a disease group and a non-disease group. (Source: Lam and Law. J Proteome Res. 2014)
Untargeted Profiling Detects Natural Antibiotics in Grassland Plants
Dr. Katherine French, a biologist from the Plant and Microbiology Department at the University of California, Berkeley and Prof. James McCullagh, Director of the Mass Spectrometry Research Facility at the University of Oxford, are no strangers to metabolomics, but in their case, the main subjects of study are plants, rather than humans. Their untargeted profiling of 17 species of wild grassland plants from livestock grazing fields detected 16,000 plant compounds, including 32 with known antimicrobial properties.
“The most interesting thing we found was that plants produce a variety of antibiotic and anthelmintic compounds that target everything from pathogen physiology, to reproduction,” French told us. “Take meadowsweet (Filipendula ulmaria) for example. It contains carvacrol, which can destroy bacterial cell membranes, and rosmarinic acid, which can inhibit biofilm formation.” French went on to say that some compounds, like tannins, have multiple functionalities: they can cause skin lesions and reduced motility in adult parasitic worms, while also inhibiting larval development.

Figure 3. Heat map of 31 antimicrobial and anthelmintic compounds found across 17 grassland plants. These compounds perform a diverse array of functions, including targeting microbial cell membrane integrity and conjugation in bacteria, disrupting quorum sensing, and reducing the motility and fertility of intestinal worms. Samples from each plant are identified as the first four letters of the genus and first three letters of the species according to Linnaean classification. (Source: French et al. Sci. Rep. 2018)
Another important finding from French and McCullagh’s untargeted metabolomics work was that plants closely related to each other from an evolutionary perspective have similar metabolic profiles. Such evidence for the genetic basis of plant secondary metabolism is extremely useful for future work in the search and investigation of plants with specific medicinal properties. “We found that plants from the Fabaceae family (the legume family, which includes plants like clover, lucerne, and soy) had a greater diversity and higher abundance of antimicrobial compounds than many others,” noted French.
These findings have far-reaching practical implications. Many of the legumes tested in the study are found naturally in meadows where livestock graze in the UK. However, they are not commonly used in conventional (non-organic) agricultural systems. Armed with extensive metabolomics data, French was able to advise farmers and seed companies on what plants to include in livestock grazing systems, in order to control microbial infections. “These recommendations can also be used by farmers outside of the UK; instead of sowing legumes native to the UK, they could sow legumes native to where they live, e.g. in Brazil or South America,” she remarked. “This would promote livestock health and maybe even contribute to local biodiversity conservation.”
As for the technologies involved, French and McCullagh chose an LC-MS/MS platform combining reversed-phase chromatography with high resolution mass spectrometry. “We were interested in obtaining as much information as possible about the small molecule profile and looking for correlations between plant species and land management; in addition, we wanted to identify natural anti-microbial compounds and how these were represented across the sample,” said McCullagh. “Using an untargeted LC-MS/MS workflow, with integrated analysis of authentic standards from an in-house library, enabled us to use a number of orthogonal measurement parameters including accurate mass, retention time and isotope pattern matching, to confidently identify selected metabolites. “
Comparing the technique to NMR, French pointed out that apart from being more sensitive, LC-MS is often easier on the research budget as well – something that needs to be considered when you have hundreds of samples to analyze. “As long as you have a good in-house database, you can identify hundreds of metabolites in your samples,” said McCullagh. For their research, French and McCullagh utilized a library of over 500 authentic metabolite standards (including over 100 plant secondary metabolites) but they also made use of online databases such as the Human Metabolome Database (HMDB) containing many plant-derived primary and secondary metabolites.
Workflow and Challenges
The workflow used for untargeted metabolic profiling starts with sample preparation. When dealing with plant specimens, freeze-drying samples for 2-3 days is essential to remove moisture without the loss of metabolites. Then, samples were reduced to a fine powder through blending and centrifuging. “By breaking down the plant material, you are making the metabolites found in the cells more accessible to the solvents used in metabolite extraction,” elaborated French. “For LC-MS/MS, we used ethanol to extract the metabolites followed by centrifugation to removed particulates. This extracts a wide range of plant secondary metabolites which were the compound classes of interest.”
“After filtration, the ethanol extracts were analyzed using the LC-MS/MS platform,” explained McCullagh. “With a combination of accurate mass analysis, retention time, MS/MS peak matching, and isotope patterns, metabolites could be compared between samples. We identified a number of these by the co-analysis of authentic standards.” Data was processed using offline and online platforms including Progenesis QI, MetaboAnalyst, and R.
The greatest challenge French and McCullagh and her team encountered during these profiling studies was the detection of hundreds upon hundreds of compounds that were completely unknown. “We couldn’t identify these compounds by using just LC/MS and publicly available databases,” she said. “Online databases of plant metabolites are far less extensively developed than for mammalian metabolism, leaving us with the majority of compounds well characterized, but not identified,” added James.
With such a wealth of information obtained via untargeted metabolomics, a next step for French and McCullagh would be to look for new anti-microbial compounds in their wild grasses. “In order to do this, we would need to split the samples into fractions, ideally containing individual compounds, and conduct antimicrobial assays to identify whether they show bioactivity,” she concluded.



