Structure of Tau Filaments from Alzheimer’s Brain Solved by Cryo-EM

Complete the form below to unlock access to ALL audio articles.
Researchers from the Medical Research Council's Laboratory of Molecular Biology, Cambridge, UK, have solved the structure of the dementia-causing ‘tau’ protein in unprecedented detail. Using brain tissue from a deceased 74-year-old Alzheimer’s patient, researchers isolated tau proteins and explored their structure using Cryo-Electron Microscopy (Cryo-EM). Their insight into how tau proteins assemble into their filaments could enable better, more definitive diagnosis of the type of dementia suffered and enable targeted treatment of the disease.
Learn more: Cryo-EM explained by Sjors Scheres
Tau proteins: Tiny but destructive
Six types of tau proteins exist and they range from 352 to 441 amino acids in length. When functioning normally in neuronal brain cells, these proteins bind to microtubules to stabilize their formation and maintenance and are thought to be protective in the brain. However, if tau protein filaments aggregate within neurons they can pile up into large tangles which disrupt the cell’s shape and affect its ability to function properly, eventually killing it. This type of cell loss causes the destructive degeneration seen in the brains of dementia patients and underlies the symptoms of the disease.
What causes the tau proteins to assemble incorrectly is still unknown. Evidence is gathering that some abnormal tau molecules with specific repeat motifs can act to influence or seed the disease in normal tau proteins and this disruption can spread, giving them infectious properties.
As senior author, Michel Goedert explains, “We have known for almost three decades that the abnormal assembly of tau protein into filaments is a defining characteristic of Alzheimer's disease. In 1998, the dysfunction of tau protein was shown to be sufficient for neurodegeneration and dementia. In 2009, the prion-like properties of assembled tau were identified. These properties allow the abnormal form to convert previously normal forms.”
Abundant tau proteins are the hallmark of tauopathies like Alzheimer’s Disease, Chronic Traumatic Encephalopathy, Pick’s disease and others. Although Alzheimer’s is slightly different in its pathology as sufferers also have characteristic plaques of amyloid beta protein in the tissue surrounding their brain cells.
Understanding how abnormal tau proteins assemble into their filaments which accumulate into fibrils is of great importance, as disrupting the formation of dangerous aggregates could facilitate the development of a treatment for tauopathies and dementia.
Read similar: tau protein linked to Alzheimer's decline
Cryo-EM: Providing insights straight from the brain
Until now, efforts to solve the structure of tau protein have needed much larger amounts of protein than those produced in a diseased brain. This meant the proteins were made using cells in a lab and the filaments were artificially made to aggregate in a dish. Although powerful insights were gained, the question of whether the tau filaments aggregate like this in the human brain remained.
One of the key benefits of utilizing Cryo-EM in this study was that less protein was required, meaning tau could be isolated directly from a diseased human brain. As lead Author, Sjors Schere’s explains, “Solving the structure of these proteins at the atomic level from actual brain tissue, for the first time, is one of the biggest accomplishments of this study.”  Structure of tau filaments from Alzheimer's brain. Left: Coronal section of brain tissue from which the tau proteins were isolated. Visible are the enlarged ventricles (grey) characteristic of Alzheimer's pathology. Right: negatively stained electron micrograph of purified tau filaments. Arrows point to paired-helical filaments (blue arrows) and straight filaments (green arrow). Credit: Scheres Group, MRC-LMB.
Structure of tau filaments from Alzheimer's brain. Left: Coronal section of brain tissue from which the tau proteins were isolated. Visible are the enlarged ventricles (grey) characteristic of Alzheimer's pathology. Right: negatively stained electron micrograph of purified tau filaments. Arrows point to paired-helical filaments (blue arrows) and straight filaments (green arrow). Credit: Scheres Group, MRC-LMB.
Resolution revolution: improved solving power
Driven by improved electron detection and better software for modeling protein structures, the Cryo-EM resolution revolution is enabling the structures of large, complex and previously unsolvable protein structures to be resolved at the atomic level. The Scheres group resolved the tau protein structure at a resolution of 3.4 - 3.5 angstroms, where an angstrom is 1 ten-billionth of a meter in size.The scientists were then able to model the assembly of the tau proteins into their filaments by using an add-on to the ‘RELION’ structural modeling software package, originally developed by Sjors’ group, written by study author Shaode He.
Filamentous findings
When the group isolated and imaged the tau filaments from the Alzheimer’s brain, they found the tau proteins were arranged in two types of filament, paired-helical filaments and straight filaments. The team mapped the structural arrangement of atoms in the abnormal tau proteins, noting they have similar structural motifs to those of amyloid and prion proteins. Prions are known to be infectious and cause scrapie and bovine spongiform encephalopathy (BSE) in sheep and cows, and Creutzfeldt-Jakob disease in humans (CJD).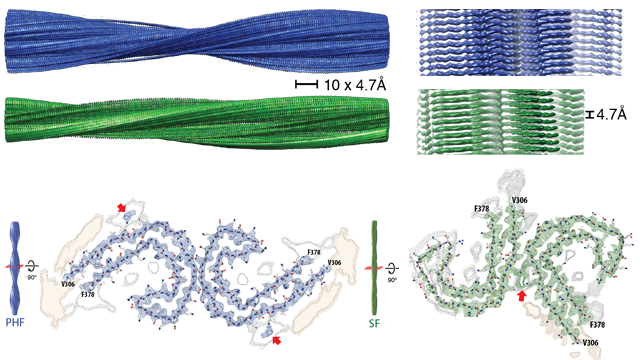 Paired-helical filaments and straight filaments. Top: Paired helical filaments (blue) and straight filaments (green). Bottom: cross section through the filaments reveals the arrangement of the tau proteins. Red arrows indicate additional protein densities that contact the tau proteins. Credit: Scheres Group MRC-LMB.
Paired-helical filaments and straight filaments. Top: Paired helical filaments (blue) and straight filaments (green). Bottom: cross section through the filaments reveals the arrangement of the tau proteins. Red arrows indicate additional protein densities that contact the tau proteins. Credit: Scheres Group MRC-LMB.
The authors show that in paired-helical filaments the arrangement of the C-shaped tau subunits is more symmetrical than in the straight filaments. However, for both paired-helical and straight filaments, the amino acids of the tau proteins that make up the core of the filamentous structure are the same. Crucially, they identified amino acid residues 306-378 as the important ‘core’ region of the proteins.
A twist in the tale for drug discovery
Sjors explains the importance of these findings for scientists working in dementia research, “The important thing for scientists working in the tau field is that we now know which amino acids are important for forming the structured core. Which means that we now know the amino acids crucial for forming this seed for aggregation of the longest forms of tau, in the diseased brain.”Sjors also highlights an issue raised by their findings for the pharmaceutical companies that recreate tau aggregation in a dish to design drugs to disrupt it. He says, “in vitro aggregation studies use tau constructs that start at amino acid residue 244 and end at 372. Meaning their constructs stop halfway through the 8th beta strand of the tau protein.” This would mean they are missing a large chunk of the core region which the group identified as key for filament formation.
Adding, “based on our findings, pharmaceutical companies should increase the length of the constructs if they want to recapitulate the formation of filaments and really test drugs to inhibit paired-helical and straight filament formation.”
Diagnosing and treating dementia
In the UK alone, 517,426 people have a dementia diagnosis, but it is estimated that the number living with the disease is much higher, at 850,000.Globally, the number of people diagnosed with dementia is increasing every year at an alarming rate. There are currently over 46.8 million people living with dementia and this is estimated to rise to 131.5 million people by 2050.
Currently, dementia is the only condition in the top 10 causes of death world-wide without a treatment to prevent, cure or slow its progression.
The insight gained from this study could improve the diagnosis of dementia. The design of ligands that bind tau and can be seen under a PET scanner to map disease progression could be made specifically to bind to the different isoforms of tau that underlie the different tauopathies. Meaning different dementias could be identified and monitored by a live imaging brain scan.
See: measuring disease progression by PET scan
Dr. Rosa Sancho, Head of Research at Alzheimer’s Research UK explains the importance of solving the structure of tau in terms of finding a cure for dementia, “A build-up of tau protein is a feature of a number of different degenerative brain diseases, and this work could help to answer key questions about why it starts to behave unusually and how it leads to such a wide variety of conditions. As well as improving our understanding of diseases like Alzheimer’s, knowing the precise structure of tau will help inform research into new treatments. Drugs that could clear away clumps of protein in the brain are a key goal for researchers, but to directly affect these proteins, molecules that make up a drug need to latch on and bind to their surface. Knowing the precise shape of these complex protein structures is enormously valuable in guiding the development of targeted drugs.”
Solving the structure of the pathological forms of tau from a human brain and the insight of how these proteins assemble into filaments is a massive boon for the tau field and will hopefully unlock the gates to better treatments and a possible cure for Alzheimer’s Disease.
And Sjors identifies the next steps for his lab, “We would now like to look at tau from brains diagnosed with other tauopathies so that we can solve the filament structures of all human tauopathies.”
You can follow @SjorsScheres on Twitter and follow his Lab's work here.




